MedTech
From the patient to the sequence
With an increased demand for rapid and accurate methods of detecting pathogenic bacteria, viruses, and other disease-causing agents, there have been significant efforts to take PCR out of the laboratory and into the field.
The field of molecular biology would not have advanced to where it is now, if was not for Kary Mullis and his Nobel prize-winning polymerase chain reaction (PCR), an essential technology for the detection and analysis of nucleic acids. Since its introduction to the scientific world in 1983, PCR has become the go-to method for amplifying specific sequences of DNA (and sometimes RNA) for a variety of purposes. Whether it is used for gene-mapping, DNA fingerprinting, detecting bacterial/viral infections, or studying genetic disorders, PCR is usually at the crux of many genomic applications.
For molecular biologists, the allure of PCR lies in its sensitivity and specificity. Most quantitative, real-time PCR assays available nowadays are more sensitive and more specific than plate-based immunoassays – they could detect down to a single nucleic acid molecule.
Digital PCR (dPCR), finding increasing popularity, partitions one sample into a large number of micro-wells, or droplets, and the reaction is carried out in each individually. This provides a collection of yes/no results and a more reliable and precise quantification. dPCR techniques are extremely useful for detecting variations like copy number variants and point mutations and has been shown to demonstrate high reproducibility, even across multiple laboratories without calibration. It overcomes the problems of background noise and relative measurements seen with qPCR (digital PCR allows absolute quantification), and enables high-throughput analysis.
From detecting cancer to microbial infections, and now even fungal infections, PCR has become the gold standard of analyzing gene expression. Due to an increased demand for rapid and accurate methods of detecting pathogenic bacteria, viruses, and other disease-causing agents, there have been significant efforts to take PCR out of the laboratory and into the field.
There are a lot of criteria to meet for field diagnostics to be successful. Accuracy, affordability, simplicity, and robustness are just some of the key criteria needed for successful PCR in remote regions. The difficulties do not stop here; another issue arises when additional equipment is required to detect the amplification product, when there are no classic lab benches or electrical outlets nearby. This also highlights the issue of high energy consumption, which can limit the use of PCR outside of the lab. To amplify DNA in remote locations, alternative methods have been developed without thermal cycles, but these alternative methods often fall short on sensitivity and require expensive reagents.
Cas9nAR. A team of researchers from the East China University of Science & Technology (Shanghai, China) has developed an amplification method using an altered version of Cas9. The method, Cas9 nickase-based amplification reaction (Cas9nAR), can occur at a constant temperature of 37°C (98.6°F) in a single step and is said to be inexpensive to perform.
Cas9 was adjusted to become a nickase so that it would only nick DNA by cutting through one strand, as opposed to completely cutting both strands. The Cas9 nickase binds to an RNA sequence that recognizes a specific DNA sequence for the site of the nick, and then proceeds to nick the DNA that is immediately adjacent.
In the new method, two different Cas9 nickase RNA complexes nick the DNA in two different places. A polymerase then complements the cut strand, starting at the first nick and moving toward the second nick, setting the old DNA strand free as it moves. The new DNA is then nicked and complemented by the Cas9 nickase complex. The short single DNA strands that are released by this process become the starting point for further amplification in the second cycle. As well as the nickase and polymerase, the only other requirements for this method are two suitable primers.
In order to confirm that the correct DNA sequence was identified, tests were performed with a fragment of bacterial genomic DNA. Furthermore, considering that the reagents can be lyophilized, the above features of Cas9nAR offer great potential to facilitate efficient nucleic acid detection in the point-of-care (PoC) settings.
COVID and PCR
At present, PCR and antibody testing are the dominant ways that global healthcare systems are testing citizens for COVID-19. Both techniques have their caveats, and as the crisis unfolds, researchers are looking into alternative ways to screen for the deadly disease.
Over the course of the current COVID-19 crisis, the importance of reliable, accessible testing to screen for the disease has become increasingly apparent.
The majority of tests for COVID-19 can be divided into PCR or serologic tests. Both of these tests use different kinds of samples to search for different hallmarks of the SARS-CoV-2 virus – and neither of them is exactly perfect.
At the moment, the majority of the current COVID-19 tests that all the reports are coming from are using PCR. They detect the genetic information of the virus, the RNA. That is only possible if the virus is there and someone is actively infected.
PCR gives us a good indication of who is infected. That is the true advantage of the current major diagnostic tests – one can break that transmission chain and get a clearer picture of what is happening.
By scaling PCR testing to screen vast swathes of nasopharyngeal swab samples from within a population, public health officials get a clearer picture of the spread of COVID-19.
However, PCR tests can be very labor-intensive, with several stages at which errors may occur between sampling and analysis. False-negatives can occur up to 30 percent of the time with different PCR tests, meaning they are more useful for confirming the presence of an infection than giving a patient the all-clear.
 During the course of the outbreak, the PCR testing has been refined from the initial testing procedures and with the addition of greater automation to reduce errors. As such, now it has an 80–85 percent specificity in detecting the virus.
During the course of the outbreak, the PCR testing has been refined from the initial testing procedures and with the addition of greater automation to reduce errors. As such, now it has an 80–85 percent specificity in detecting the virus.
Recently, new testing guidelines for COVID-19 issued by the Indian Council of Medical Research (ICMR) has also allowed rapid antibody testing in India which help the agencies to enhance screening and identify capabilities. The rapid testing kits are quicker to get early results.
Instead of detecting viral genetic material, rapid tests kits target the immune response of the person being infected, looking out specifically for antibodies against the virus or virus antigens. The trouble is that antibodies only develop several weeks after an infection, which means that antibody-based tests might miss asymptomatic cases or people in the earliest stage of the disease.
That is not to say that antibody-based tests are not useful. On the contrary, antibody-based tests have proven to be crucial in linking clusters of infection, by detecting people who were infected but discovered too late to test positive via RT-PCR. As RT-PCR looks for virus RNA, it will only give a positive test result if there is an ongoing infection. On the other hand, antibodies can persist for months or years, allowing tests to identify anyone who has ever been infected.
Indian market
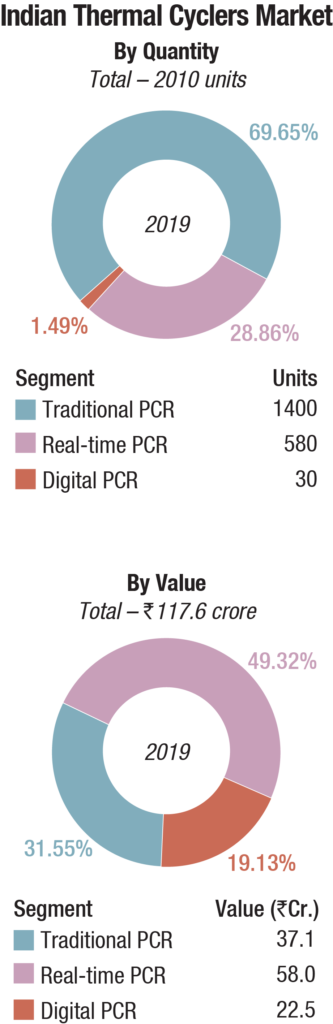
The Indian market for thermal cyclers, catering to the life sciences segment in 2019, is estimated at Rs 117.6 crore. The traditional PCRs dominate the segment, by volume with a 69.65-percent share, and have a 31.55-percent share, by value. The real-time PCRs dominate the segment by value with a 49.32-percent share, and have a 28.86-percent share, by volume. The digital PCR, which was introduced in India around 2016, is the fastest-growing segment. Its share by value is 19.1 percent and by volume 1.49 percent. The traditional segment is gradually being eased out, in preference to the real-time PCRs, and some brands are offering a complimentary one with every real-time sold.
Thermo Fisher and Bio-rad dominate the segment. Eppendorf and HiMedia are neck-to-neck in the traditional PCR segment as are Roche and Agilent in the real-time category. Agilent, Analytik Jena, and some Taiwanese, Korean, and Chinese companies also have presence. Whereas Agilent and Analytik Jena are restricted to the northern part of the country, competitive pricing has almost forced out the East-Asian brands, represented by the smaller distributors.
Digital PCR (dPCR) is finding great acceptance in India. It is used for liquid biopsies in the areas of cancer diagnosis, monitoring, evaluation of treatment modalities, and testing for reoccurrence after remission. Diabetes and organ transplantation monitoring are discussed to demonstrate other emerging uses for liquid biopsy. It is a powerful technology that provides absolute quantification of nucleic acids with a high degree of sensitivity and precision.
In blood, the cfDNA, ctDNA, and CTCs of interest are present at low levels and found in a complex background of other components. Additionally, circulating DNA is highly fragmented, which further reduces the concentration of intact target sequence. Digital PCR has permitted the detection and quantification of low-abundance targets in shorter times without requiring large numbers of replicates. This has established dPCR as a tool of choice for liquid biopsies.
The multiplex PCR method is now being used for amplification of a genome of more than 30 kb. This protocol opens possibilities in the field of molecular virology, firstly because it can be easily applied to sequence and assemble other virus genomes. It is also of particular interest to explore over time the evolution of a virus genome during the infection process as one can follow which variants take over the others.
The Union Health Ministry recently announced that the reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard frontline test for coronavirus (COVID-19) and that rapid antibody test cannot replace it. The statement came after the ICMR’s national taskforce issued guidelines on testing strategy to all states, following a review of the worldwide testing methodology.
Presently, the government is using the RT-PCR tests to detect novel coronavirus from samples of throat or nasal swabs of people with symptoms, or high-risk individuals who might have come in contact with positive patients.
What is real-time RT-PCR? Real-time RT-PCR is a nuclear-derived method for detecting the presence of specific genetic material from any pathogen, including a virus. Originally, the method used radioactive isotope markers to detect targeted genetic materials, but subsequent refining has led to the replacement of the isotopic labelling with special markers, most frequently fluorescent dyes. With this technique, scientists can see the results almost immediately while the process is still ongoing; conventional RT-PCR only provides results at the end.
Leading Players in Indian Thermal Cyclers Market |
||||
|---|---|---|---|---|
Major players* – 2019 |
||||
| Segment | Tier I | Tier II | Tier III | Others |
| Traditional PCR | Thermo Fisher | Bio-Rad | HiMedia & Eppendorf | Agilent, Analytik Jena (Biometra), Unorganized, Taiwanese & Korean brands |
| Real-time PCR | Thermo Fisher | Bio-Rad | Roche & Agilent | HiMedia, Analytik Jena (Biometra), & Chinese brands |
| Digital PCR | Bio-Rad | Thermo Fisher | – | – |
| *Vendors are placed in different tiers on the basis of their sales contribution to the overall revenues of the Indian thermal cyclers market. | ||||
| ADI Media Research | ||||
Viruses as the coronavirus (SARS-Cov2) only contain RNA, which means they rely on infiltrating healthy cells to multiply and survive. Once inside the cell, the virus uses its own genetic code – RNA in the case of the coronavirus – to take control of and reprogram the cells so that they become virus-making factories.
In order for the coronavirus to be detected early in the body, using real-time RT-PCR, scientists need to convert the RNA to DNA. This is a process called reverse transcription. They do this because only DNA can be copied – or amplified – which is a key part of the real-time RT-PCR process for detecting viruses.
Scientists amplify a specific part of the transcribed viral DNA hundreds of thousands of times. A sample is collected from parts of the body where the coronavirus gathers, such as a person’s nose or throat. The sample is treated with several chemical solutions that remove substances, such as proteins and fats, and extracts only the RNA present in the sample. This extracted RNA is a mix of a person’s own genetic material and, if present, the coronavirus’ RNA. The RNA is reverse transcribed to DNA, using a specific enzyme. Scientists then add additional short fragments of DNA that are complementary to specific parts of the transcribed viral DNA. These fragments attach themselves to target sections of the viral DNA, if the virus is present in a sample. Some of the added genetic fragments are for building DNA strands during amplification, while the others are for building the DNA and adding marker labels to the strands, which are then used to detect the virus.
The mixture is then placed in a RT-PCR machine. The machine cycles through temperatures that heat and cool the mixture to trigger specific chemical reactions that create new, identical copies of the target sections of viral DNA. The cycle repeats over and over to continue copying the target sections of viral DNA. As new copies of the viral DNA sections are built, the marker labels attach to the DNA strands and then release a fluorescent dye, which is measured by the machine’s computer and presented in real time on the screen. The computer tracks the amount of fluorescence in the sample after each cycle. When the amount goes over a certain level of fluorescence, this confirms that the virus is present. Scientists also monitor how many cycles it takes to reach this level in order to estimate the severity of the infection – the fewer the cycles, the more severe the viral infection is.
Why use real-time RT-PCR? The real-time RT-PCR technique is highly sensitive and specific, and can deliver a reliable diagnosis as fast as three hours, though usually laboratories take on an average between 6 and 8 hours. Compared to other available virus-isolation methods, real-time RT-PCR is significantly faster and has a lower potential for contamination or errors as the entire process can be done within a closed tube. It continues to be the most accurate method available for detection of the coronavirus.
Global market
The global real-time PCR and digital PCR market is estimated to grow to USD 6270.9 million by 2024 from USD 4113.3 million in 2019, at a CAGR of 8.8 percent, predicts Research and Markets. High prevalence of target diseases and rapid technological advancements such as development of high performance and superior qPCR and dPCR systems are expected to be the key factors driving the growth.
Continued efforts by PCR manufacturers to develop novel systems and kits to use in molecular testing are likely to further propel growth of the qPCR and dPCR market. In addition, a broad range of application of qPCR and dPCR technologies is expected to further boost the adoption in the array of scientific disciplines. The technologies have wide-ranging applications in both basic and diagnostic research. They have been extensively used in areas, such as human genetic testing, forensic sciences, pathogen detection, and infectious disease testing. Expanding application in food microbiology and veterinary medicine among other industries is expected to further fuel the market.
Technology insights. Quantitative PCR held the largest market share in terms of value in 2019. Continued demand in areas such as genetic variation analysis, gene expression analysis, and genotyping, along with increasing focus on the development of PoC platforms, is anticipated to drive the segment. Digital PCR is a next-generation testing method, which helps precise quantification of nucleic acids. These systems offer superior sensitivity and accuracy compared to other PCR-based approaches, which is anticipated to help the segment register robust growth in coming years.
Product insights. The consumables and reagents were the leading segment accounting for the largest market share in terms of value in 2019. The segment is projected to witness strong growth rate as compared to other segments owing to launch of new PCR test kits.
Application insights. Research application was the dominant segment in 2019. qPCR and dPCR systems are widely used for research applications such as stem-cell research and genetic disease and oncology research. The wide-spread use of these systems is attributed to features like enhanced specificity and quicker turnaround time. The clinical application segment is estimated to register the fastest CAGR of 8.7 percent with the increasing adoption of automated processes.
Regional insights. North America led the regional segment in 2019, attributed to launch of new systems and test kits by the PCR manufacturers. Moreover, the strong presence of major PCR manufacturers in this region, coupled with increasing demand for rapid diagnostic tests, is also expected to drive the regional growth over the forecast period.
Asia-Pacific is likely to witness the fastest CAGR of 11.1 percent. Rise in the patient pool owing to high prevalence of chronic and infectious diseases, coupled with increasing awareness among patients about early diagnosis of these diseases, is anticipated to boost the demand for PCR products, in turn driving the growth of the market.
Market share insights. Major players have been consistently focusing on deploying strategic business measures to maintain their competitive position. The key measures adopted by the players include new product launches, entering into collaborations and partnerships with institutes and companies, and mergers and acquisitions.
For instance, in January 2019, Qiagen N.V. announced the acquisition of Formulatrix’s digital PCR assets in a deal valued at USD 260 million. The acquisition augers well with the company’s strategy of launching new digital PCR platforms in 2020 by incorporating Formulatrix’s advanced dPCR technology.
Some of the prominent global players that dominate the market for qPCR and dPCR are Bio-Rad Laboratories, Thermo Fisher Scientific, Qiagen N.V., Abbott Laboratories, Inc., Agilent Technologies, Inc., bioMerieux S.A, Fluidigm Corporation, and Roche.
Future PCR
The diagnostic and clinical demands for PCR are increasing. Digitization of PCR is one way to address the needs that PCR alone cannot. Droplet Digital PCR (ddPCR) is a method for performing digital PCR that is based on water-oil emulsion droplet technology.
What makes ddPCR poised to address clinical needs where other types of PCR cannot? Whereas qPCR requires a standard curve for absolute quantification, ddPCR does not. It thus enables determination of blood biomarker concentrations (or ratios of multiple markers), above and below which clinical decisions should be made. Unlike qPCR, ddPCR also demonstrates both inter- and intra-lab reproducibility.
For SNVs in particular, which comprise a large fraction of the gene alterations driving cancer, ddPCR enables greater sensitivity for rare mutant allele detection. This enables powerful applications such as the early detection of treatment response and disease recurrence or progression through liquid biopsy. The ddPCR technology can also work well with other established technologies to address clinical needs. For example, next-generation sequencing (NGS) and ddPCR can work together to identify signs of cancer in liquid biopsies: NGS provides a panoramic view of genetic mutations associated with cancer, and ddPCR technology enables serial monitoring of these key mutations.
Way forward
More than 30 years after the invention of PCR, it is rare to find a molecular biologist that has not used this technology. Now, thanks to approaches like ddPCR, miniaturized instruments, and nucleic acid amplification and analysis, biologists have a new insight into rapid and accurate quantitative diagnostics for the future PoC applications.
Recent examples only highlight a fraction of the improvements made to this old technology. Each time a question is asked of PCR, researchers have found a way to make it work; from remote sub-Saharan Africa to onboard the International Space Station, no task seems too difficult.
But will the clinical demands eventually exceed the capability of this technology?
Second Opinion
 Dr Jyoti Upadhyay
Dr Jyoti Upadhyay
Assistant Professor,
School of Health Sciences, UPES
A versatile technology
Thermal cycler or DNA amplifier or PCR machine has become a standard tool in biomedical research globally. Earlier, the PCR reaction was a time-consuming process. Advancement in the newer technologies generates a more precise, time-saving, all-in-one machine, a thermal cycler that automates the PCR process and advances difference analysis, gene knockout technology, gene delivery, gene therapy, cloning of known and novel genomic DNA, cDNA sequences, quantification of mRNA and DNA, and construction of chimeric or mutant DNAs. Development of the fast PCR system has speeded up the thermal cycler run times from almost two hours to less than forty minutes. Before that, the PCR was limited to only 4 runs per 8 hours a day. Another advancement in the fast PCR system is the ability to achieve faster temperature ramp rates and the completion of a successful PCR reaction of thirty cycles in fifteen minutes.
Advancement in the design of thermal cyclers offers faster processing and protocols. They have undergone advancements in their mechanics like lid security, design specifications, and heal-block reliability over the past few decades. Thermal cycler with heat blocks 8 to 384 wells are available to accommodate 200 microliter PCR tubes. Researchers have the options to choose dual and multi-block PCR instruments, with optimizing genotyping productivity. Miniaturized thermal cyclers have a microfluidic chip, in which the tubes containing reaction mixtures move through hot and cold zones. Modern thermal cyclers are equipped with fully adjustable heated lids that allow the use of diverse types of PCR plastic ware.
Thermal cyclers or PCRs are used as routine technique in various laboratories, allowing analysis of very small amounts of DNA or damaged DNA samples. They are commonly used for a wide variety of applications like in diagnosing genetic diseases, formation of a genetic fingerprint (also known as DNA profiling) from a blood or semen sample, or from a hair root; diagnosis of infectious diseases (detect the presence of antibodies); personalized medicine to select patients for certain treatments and to create copies of DNA for introduction into host organisms such as Escherichia coli in genetic engineering, and to amplify stretches of genetic material for Sanger sequencing – the Human Genome Project used PCR.
 Dr Tasleem Raza
Dr Tasleem Raza
Professor
Eras Lucknow Medical College
PCR, a gold standard
Since the discovery of the polymerase chain reaction (PCR) in 1983 by Kary Mullis, an avalanche of scientific publications has reported major developments in specialized equipment, reagents, sample preparations, scientific techniques, and university research and hence this technique has become a revolutionary watershed for medicine and science.
It is nearly a standard and reliable technology, which is used to amplify and produce a million copies of a single molecule of DNA or RNA for genotyping, cloning, and analysis of single nucleotide variations of etiologic significance.
The healthcare landscape has also benefitted from the advancements in quantitative polymerase chain reaction, or qPCR first described in 1992, which has become the basis of a variety of scientific applications and publications in a broad range of interests. It allows a precise and rapid detection of nucleic acid by incorporates intercalating dyes or sequence-specific probes for fluorescence into nascent DNA.
On the other hand, the reverse transcription-PCR (RT-PCR) assays have substantially accelerated the momentum and accuracy of human and animal disease diagnosis, especially the infectious agents that were earlier difficult to isolate or demonstrate. It addresses the evident need for quantitative data analysis and snapshots the information regarding the quantity of a given transcript in a cell or tissue.
Any discussion regarding the advances in PCR would be unworthy with a mere mention of dPCR (digital PCR), which is recent technology available commercially since 2011, which utilizes pre-validated primer or probe assays through partitioning of a PCR reaction into hundreds or thousands of individual vessels, typically in droplets or wells with the acquisition of data-positive or negative for every droplet, allowing greater amenability and multiplexed detection of target molecules.
Genotyping studies allow the study of bacteria like Mycobacterium tuberculosis which is of tremendous worth for public health, favoring the early recognition and optimized treatment. Thus PCR is a gold standard technique to genetically characterize any microbial isolate and facilitate its epidemiological studies.