DNA Sequencers
India pays a heavy price for neglecting genome sequencing

The genome sequencing exercise, running at a snail’s pace, is expected to gather momentum as the authorities realize the challenges that insufficient sequencing and pathogen surveillance pose.
The emergence of new variants of the novel coronavirus (SARS-CoV-2) have set off alarm bells across the world, including India. However, India’s genome sequencing program has a long way to go. As researchers stress the importance of genome sequencing to gauge the impact of the new mutant variants, three of which have invited concern so far, the scale of exercise in India has been dismal.
The number of COVID-19 cases in India has just crossed the 25-million or the 2.5 crore mark. That is a 16 million increase from November 23 — the date from which the government said it would do the genome sequencing of 5 percent of positive cases. Which means that by now, the Indian SARS-CoV-2 Genomic Consortium (INSACOG) should have sequenced 800,000 genomes of the virus. A May 6 press statement from the Department of Biotechnology said that, so far, about 20,000 samples had been sequenced. Other countries kicked off the exercise right at the beginning of the pandemic. Given India’s significant caseload, it would have been prudent to jump on to the bandwagon quickly as well.
It was only on December 30, 2020 that the Union Health Ministry instructed the Indian SARS-COV-2 Genomics Consortium (INSACOG) to ramp its sequencing efforts. And announced that from then on, 5 percent of all positive COVID-19 samples in every state – and 100 percent of all positive samples from international travellers – will be collected on a weekly basis for sequencing. The challenge of insufficient genomic sequencing is further compounded by slow pace of data sharing.
INSACOG prominent research labs across the country include National Institute of Biomedical Genomics (NIBG), Kolkata; Institute of Life Sciences (ILS), Bhubaneswar; National Institute of Virology (NIV), Pune; National Centre for Cell Science (NCCS), Pune; Centre for Cellular & Molecular Biology (CCMB), Hyderabad; Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad; Institute For Stem Cell Science and Regenerative Medicine (INstem), Bengaluru; National Institute of Mental Health and Neuro-Sciences (NIMHANS), Bengaluru; and Institute of Genomics and Integrative Biology (IGIB), Delhi and National Centre for Disease Control (NCDC), Delhi. A decision to add 17 more laboratories, to the existing 10 labs had been announced on May 18, 2021.
According to INSACOG’s guidance document released December, these institutes have a capacity to sequence more than 30,000 samples a month. IGIB, for example, can sequence more than 10,000 samples a month. But till March 18, it had received only 3186 samples of which 1586 had been sequenced and the rest were under process.
Clearly, the SARS-CoV-2 genome sequencing program is not going as per planned.
The question becomes what is holding INSACOG from going full throttle? Several factors seem to be at play here.
First, in the initial days of the pandemic in India, the Indian Council of Medical Research had taken a considered view that sequencing was not required as the virus was stable. However, there were already reports of unique variants circulating in India before this release came out.
INSACOG still faces bureaucratic hurdles. It was initially allocated ₹115 crore for a six-month period. The money was to come from the Central government. However, no additional allocation was made and the Department of Biotechnology was asked to find the money from its own resources. The first tranche of money could be released only on March 31, the last day of the financial year. The allocation itself has been reduced to about ₹80 crore because that’s what the DBT could come up with. In the meanwhile, laboratories like the Centre for Cellular and Molecular Biology in Hyderabad, and the Institute of Genomics and Integrated Biology (IGIB) in Delhi, have been sequencing the genome using their limited funds. Labs have very limited resources. To find money to do something extra is a very difficult proposition. Most labs diverted money from other running projects to work on sequencing, in the hope that they would be compensated sometime later. Without the money, most labs cannot buy the equipment they need, and are progressing slowly with their own funds.
| INSACOG laboratories | |||
| Name of laboratory |
Capacity to sequence (Samples per month) |
Samples sequenced (January-March 18, 2021) |
Samples under process (As on March 18, 2021) |
| NCDC | 3000 | 3644 | 2791 |
| IGIB | 10,000 | 1586 | 1600 |
| NIV | – | 886 | 14 |
| CCMB | 5000 | 467 | 133 |
| NIBG | 5000 | 679 | 225 |
| NIMHANS | 1200 | 122 | 4 |
| NCBS-ISTem | 1200 | 176 | 0 |
| CDFD | 1200 | 0 | 0 |
| ILS | 1200 | 104 | 32 |
| NCCS | 1200 | 0 | 0 |
| MoHFW guidance document on genome sequencing; NCDC | |||
The funds needed are not astronomical. A small fraction of the PM CARES Fund, had it been allocated in time to some of these labs, would have enabled the scientists to strengthen their facilities, analyze the India-specific genes in detail, sequence them and bring the findings to the notice of the scientific community, so that the units manufacturing the vaccines may be in a position to fine tune the vaccines to counter the new mutants from time to time. It is a continuing effort in virology and the Indian scientific community, which can compete with the best in the world, should have unstinted support from the government in terms of funding and other facilities.
But funds were just one problem. There has also been lack of clear directives and goals.
The work is also hampered by the Union government’s order of May 2020, which stopped government labs from importing goods worth less than ₹200 crore, and general financial rules (GFR) were amended to disallow global tender enquiries. This was an attempt to protect micro, small and medium enterprises (MSMEs) from the downturn and create assured demand for their goods and services. Under the earlier guidelines for purchase, government agencies were required to procure goods and services worth less than ₹50 lakh from local suppliers. The government departments were given free hand to raise the upper limit. Exemptions could be made only if the quality or specification was not available with the local supplier. Under the purchase preference policy, if the lowest or the best bid in a government tender was from a non- Indian company, only half of the contract value was awarded to it. The lowest bidder among Indian companies would get the remaining 50 per cent of the tender, provided it matched the lowest tender price. This enabled government departments to meet the mandatory procurement goal of 25 per cent from MSMEs and a sub-target procurement of 4 per cent goods and services from MSME entrepreneurs within SC/ ST communities.
The order was meant to boost local manufacturing, in line with the Make in India initiative, but had unintended consequences for India’s fledgling genome-sequencing efforts. Several reagents and plastics used by Indian labs come from foreign manufacturers, like the US-based Illumina Incorporation, and have no Indian substitutes.
The ministry’s sudden restrictions threw these labs out of gear. By September 2020, sequencing across the country had come to a near-complete halt, as labs ran out of reagents they needed. Then, in response to their complaints, the ministry exempted reagents from its restrictions in January 2021.
But plastics materials still have not been exempted from the ministry’s order, which means labs cannot buy them in bulk unless it can conduct a market assessment to show that no Indian alternatives exist. Such an assessment is a needless bureaucratic distraction at a time when labs desperately need to sequence more viral genomes – and faster.
The collection of samples is a taxing process. According to the Union Health Ministry guidelines, samples from only RT-PCR tests — and not the more common rapid antigen tests — would be eligible for sequencing as the latter is known to sometimes result in false negatives. The samples have to be sent within three-four days of coming out positive. Otherwise the purpose is defeated. The samples are to be stored in these institutes at -80 degrees Celsius. All these 10 labs can do it. However, the problem is with logistics that pertain to samples coming from the states.
The poor progress in genome sequencing also affects India’s image abroad, as all countries are required to upload data into a common global repository, Global Initiative on Sharing all Influenza Data, or GISAID. This is to alert the world about new mutants. The world is watching apprehensively at India’s massive sweep of the second wave and expecting India to capture variants and put the data into the common database.
How much of India’s data has been put into the pool is not known, but the May 6 note said 50 percent of the data has been uploaded in GISAID. Since the samples sequenced were about 20,000, only about 10,000 have been put into GISAID. As a percentage of the total number of cases in India so far—25 million—this is a miniscule 0.004 percent.
The world is moving toward next generation sequencing. There are several NGS platforms, but the common feature is that they analyze thousands of bits of DNA chains simultaneously. The process is faster by orders of magnitude. NGS is an important tool in disease control and origin tracing. At such a time, India seems to be a laggard.
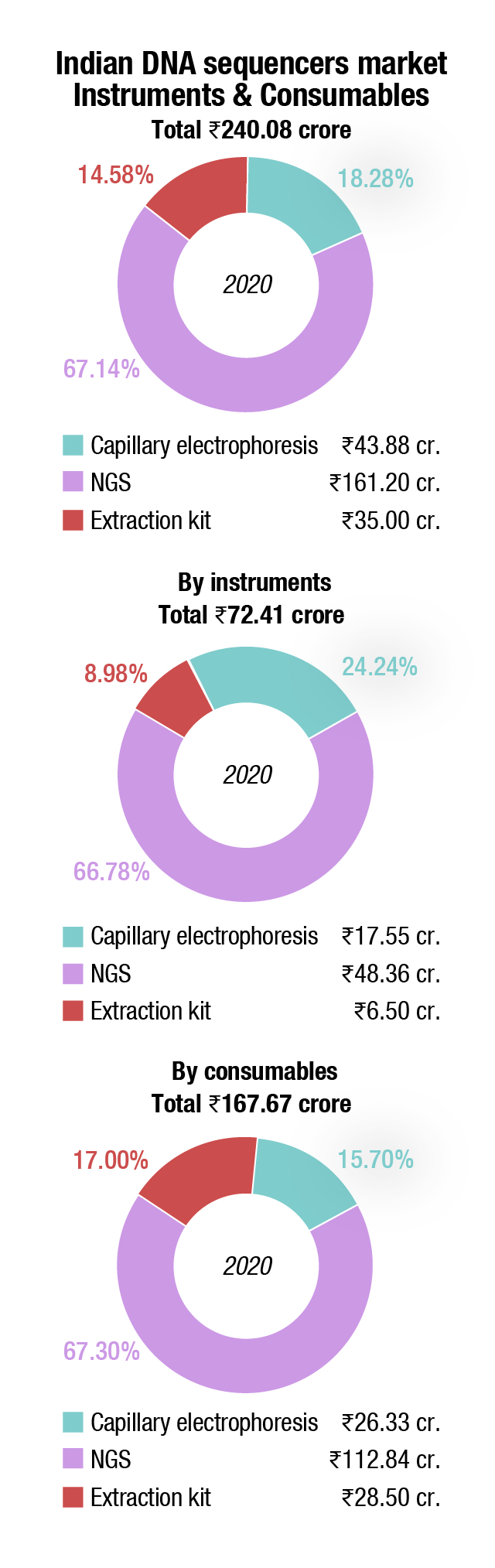
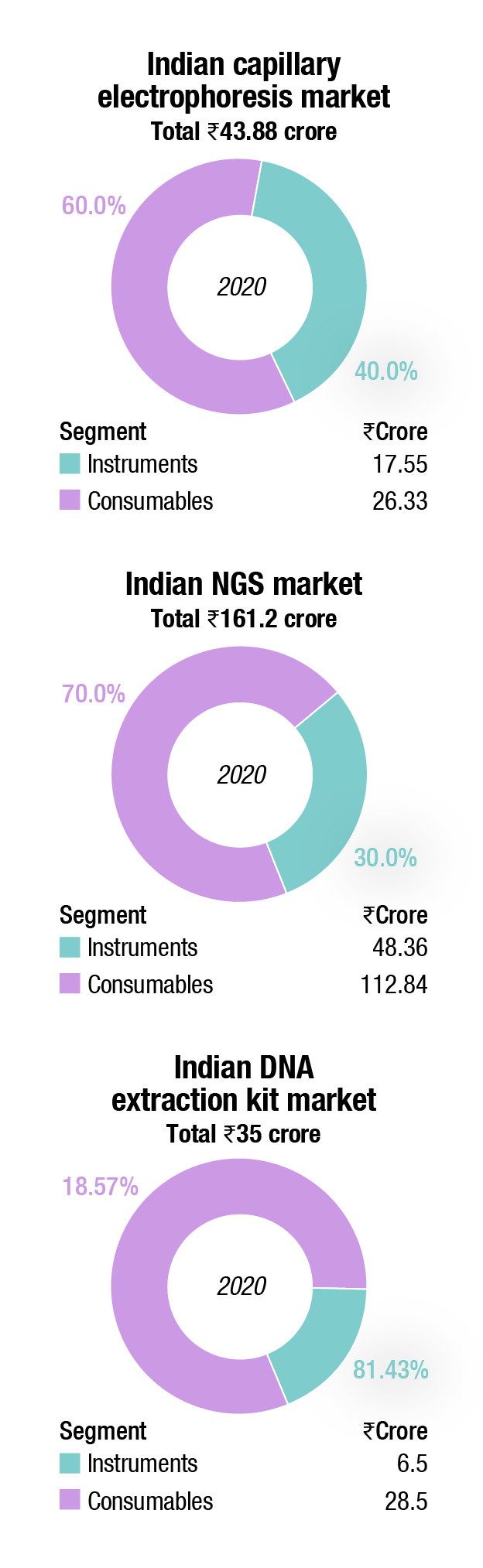
The DNA sequencers market saw a sharp drop in 2020, of 31.5 percent. With research and clinical institutes under lockdown, the technicians were moved to conducting the PCR tests.
What really took off was RNA extraction. And this trend is expected to stay. Several protocols for RNA extraction and RT-qPCR that are simpler and less expensive than prevailing methods are posible. First, isopropanol precipitation is shown to provide an effective means of RNA extraction from nasopharyngeal (NP) swab samples. Second, direct addition of NP swab samples to RT-qPCRs is evaluated without an RNA extraction step. A simple, inexpensive swab collection solution suitable for direct addition is validated using contrived swab samples. Third, an open-source master mix for RT-qPCR is described that permits detection of viral RNA in NP swab samples with a limit of detection of approximately 50 RNA copies per reaction. Quantification cycle (Cq) values for purified RNA from 30 known positive clinical samples showed a strong correlation (r2 = 0.98) between this homemade master mix and commercial TaqPath master mix. Lastly, end-point fluorescence imaging is found to provide an accurate diagnostic readout without requiring a qPCR thermocycler. Adoption of these simple, open-source methods has the potential to reduce the time and expense of COVID-19 testing.
COVID-19 has drawn attention to the relevance of sequencing and pathogen surveillance. If there is one tool in the COVID-19 pandemic response, which India has been slow in adoption and has used sub-optimally, it is genome sequencing. The scale of exercise has been dismal. To cite data, less than 1 percent of the total positive samples since January 2021 through March 18, 2021 have been sequenced.
Genome sequencing is an exercise that studies changes in the structure of the virus over time. A combination of changes (mutations) in the ribonucleic acid of the virus can give birth to a new variant. The exercise is carried out only on samples that have tested positive for the virus.
Other countries kicked off the exercise right at the beginning of the pandemic. In fact, many of its neighbours in south and southeast Asia have re-embarked on a remarkable effort to quickly scale up their very limited sequencing capacities to identify whether India’s epidemic—fuelled by variants such as B.1.617—is spilling over into their communities, or if their outbreaks have origins elsewhere. In some cases, they are working around the clock and with insufficient resources. Specifically, they want to know whether worrying variants are circulating, so that they can assess the risks. Identifying worrying variants will help governments to make decisions about responses and restrictions, and whether more aggressive interventions are needed. Given India’s significant caseload, it should have jumped on to the bandwagon quickly as well.
However, Dr V Ravi, Virologist and Chairman of Karnataka Genomic Surveillance Committee maintains that India is not lagging behind. India contributed to the world with the discovery of a very important variant. The need for sequencing arose when the Alpha variant began spreading across the globe. The Indian government acted quickly and to that extent, has not missed the bus. During the second surge there was lakhs of cases, and the amount of sequencing done was less. But India did identify that the Delta variant is one of the major ones that contributed to the surge in India.
“We are doing sequencing which at this point is based on the WHO framework of surveillance where every state has 10 sites – five hospitals and five labs. There is a clear protocol where 150 samples are to be collected every 15 days in these sites and these 10 sites will be connected to all the districts within the state. There was a delay because globally, there was an extreme shortage of reagents for sequencing. There is a huge rat race to procure. Now it is eased out since companies have begun to supply. Now onwards, regular surveillance will take place. Probably by the end of July, we will hit the five per cent mark. State governments have to pitch in and build capacity. This is a long term investment for future pandemics,” reiterates Dr Ravi.
It is expected that government will allocate more than the current 0.8 percent of GDP for this segment. In any case, the Department of Biotechnology and the Ministry of Health are looking at how to involve the private sector. Discussions are on with certain major corporate hospitals and private labs that are into sequencing. The government cannot fund them because the amount of money that is required is huge. A framework is being developed where private entities can do sequencing, but all sequences should finally find one repository in terms of data repository so that the whole nation’s picture is reflected.
Genomics is also important for other epidemic diseases like dengue, chikungunya, influenza because once COVID goes down, these will be back. Monitoring for variants and mutants there is also very important. Emerging variants, with evidence of higher transmissibility and immune escape, demand re-strategised responses.
As India prepares for the third wave, increasing genomic sequencing and use of scientific evidence for decision making are not a choice but an absolute essential.
The global DNA sequencing market is expected to grow by USD 17.92 billion during 2021–2025, as per Technavio. This marks a significant market slow down compared to the 2020 growth estimates due to the impact of the COVID-19 pandemic in the first half of 2020. However, healthy growth is expected to continue over the next 5 years, and the market is expected to grow at a CAGR of over 19 percent.
Among the product segment, consumables are expected to dominate the overall market. The wide availability of reagents and kits to cater to all steps of library construction, such as DNA fragmentation, enrichment, adapter ligation, amplification, and quality control contributes to a larger share of the segment. Most of these tools feature streamlined and simplified workflows, ready-to-use components, and compatibility with low-input and formalin-fixed specimens. The constant introduction of reagents and kits to make WGS more accessible and affordable also drives the segment in the market for DNA sequencing. For instance, in August 2020, Illumina Inc. launched NovaSeq 6000 v1.5 reagent kit, which made the WGS technique more cost-effective and accessible for all the laboratories and reduced the cost to sequence a human genome for USD 600. These new kits can be used across WGS, single-cell genomics, and liquid biopsy techniques.
The third-generation sequencing methods, such as nanopore and single-molecule real-time sequencing, are projected to witness a considerable CAGR in coming years. This technology overcomes the issues of second-generation techniques as it includes easy sample preparation without the need for PCR amplification with faster speed. Moreover, it generates long reads exceeding numerous kilobases for the detection of repetitive sites of complex genomes.
The global market for DNA sequencing witnesses high competition with Illumina, Inc.; Thermo Fisher Scientific; Agilent Technologies; QIAGEN; and Perkin Elmer as dominant players in 2020.
 Nagendra Kumar Singh,
Nagendra Kumar Singh,
National Professor BP Pal Chair,
National Research Centre on Plant Biotechnology
“DNA sequencing has now become an essential tool for rapid progress in the field of agriculture, medical and environment management. The DNA sequencing technology has gone through a sea change with rapid advances in microfluidics, nanotechnology, and informatics. With the availability of latest single molecule real time sequencing platforms there is no need to clone or even PCR amplify the DNA before sequencing. DNA isolated from any life tissues using standard methods can be used directly in the sequencing machines with minimum amount of reagents and gets not only the high sequence output but also any epigenetic modification of DNA such as methylation which have profound effect on gene expression. These innovations have greatly improved the speed, quality, and economy of DNA sequencing.
Presently there are five platforms in vogue with varying scale and economy of operation and can be utilized as per the need of the user. These are:
Sanger’s sequencing using automated capillary sequencers of Applied Biosystems (now Life Technologies). This technology helped decode hundreds of small microbial genomes and large plant and animal genomes including Arabidopsis, rice, C. elegance, and human, but now it is not used in genome sequencing due to high cost per base. However, it is suitable for less number of samples and does not require high depth of coverage due to high quality sequence output. It offers the lowest cost on per sample for sequencing small number and size of DNA fragments.
Ion Torrent platform, which is an improved and cost-effective version of now obsolete Roche 454 pyrosequencing technology. It is suitable for faster medium throughput targeting re-sequencing such as pooled amplicon sequencing of specific genes. Thousands of amplicons from hundreds of samples can be pooled after barcoding and sequenced in single run; it is great for variant analysis and DNA fingerprinting applications.
Illumina’s NovaSeq 6000 is the highest throughput sequencing platform available today. The sequence reads are short (100-150 bp) but high-quality base calls are achieved due high depth of coverage. It has the lowest cost per base pair due to high volume of data generated per run but cost per run is high therefore not suitable for low and medium throughput applications. It is used widely for genome re-sequencing for variant discovery and genotyping by sequencing
Single molecule real time (SMRT) sequencing using PacBio Sequel II system is now a preferred option for sequencing large complex genomes and transcriptome assemblies due to its high-quality long average read length of greater than 10 kbp that helps in the contig assembly. The long sequence reads have helped resolve highly heterozygous genomes by separating the maternal and paternal chromosome reads allowing phase separated assemblies. PacBio IsoSeq system enables sequencing of full-length RNA transcript and discover alternate splicing variants.
Oxford Nanopore DNA sequencing platform also generates long sequence reads of more than10 kbp, is very fast and highly cost effective. The initial high sequencing error rates of the system has been improved drastically and now it is used is increasing in rapid diagnosis of microbial samples by whole genome sequencing and even assembly of large eukaryotic genomes e.g. tomato”
Although genomics is a relatively naïve field, but over time and across the globe researchers are exploring its diverse and insightful utility. Indeed, the advancement in sequencing technologies has allowed us witness a surge in -omics terminology and their application-based studies including transcriptomics, haplomics, epigenomics, metagenomics, nutrigenomics etc. Undoubtedly, in the past one decade different methods of bulk sequencing of particular cells or tissues have immensely uncovered the underlying morphology and physiology. However, increasing evidence suggests that at times genome integrity and gene expression are heterogeneous, even in similar cell types, and this stochastic behavior reflects cell type composition and can also trigger cell fate decisions.
Thus, in order to have more resolution or understanding this cell-to-cell variability, the method of single cell sequencing has emerged. Even though this method was initially showcased in 2009, it gained real pace only in the last 5 years, spanning wide range from human to plants. This explosive growth of single cell sequencing studies is mainly attributed to simultaneous improvement in low cost sequencing and cell isolation methods. Noteworthy, this is equally supported with evolving computational algorithms for meaningful patterns and insights. As the sequencing-based research expands, now we have several derivative approaches such as single cell expression (spatial gene regulation), single cell ATAC (assessing chromatin state dynamics), single cell CNVs (copy number variants detection) etc. Moreover, single-cell multiple sequencing technique (scCOOL-seq) that allows simultaneous analysis of single-cell chromatin state/nuclear niche localization, copy number variations, ploidy and DNA methylation, which can showcase an altogether integrated view of the cell state. Currently, it is apparent that medicinal research is rapidly harnessing this single cell-based exploration, whereas plant biology is lagging little bit due to technical challenges in isolation of specific cells. However, efficient Microfluidic technology has provided some assistance. These methods of single-cell isolation has gained popularity due to its low sample consumption and low analysis cost together with the fact that it enables precise fluid control for better resolution.
 Awanindra Kumar
Awanindra Kumar
HOD-Pathology,
Swami Dayanand Hospital
“Blood is composed of solid and liquid components. Based on different relative density, sediment rate and size, a cell separator can be used to separate different blood components. Each blood component is used for a specific clinical indication; thus maximizing the utility of one whole blood unit for multiple patients. The conventional component separation has limitations in preparation of single donor components, and as per current trends it is desirable to at least have facility to prepare single donor platelet (as a lifesaving therapy in dengue viral fever) and single donor plasma (was widely used in management of COVID-19 pandemic) using cell separator. Besides this, double unit red cell collection and leucopheresis can also be prepared using same equipment, but the use of these is still to reach on significant levels. Technically, operator friendliness of equipment and ease of operation with minimum need of tech support will certainly provide an edge in the segment. Anyhow, role of blood cell separator and blood bank in coming days will be more comprehensive as single donor fractioned products is the new trend.”
Overall, experts believe that the ultimate sequencing platform would work on single DNA or RNA molecules without any (pre-) amplification and without use of optical steps. It would provide reads of Mb to Gb in length with high read accuracy and no GC bias. It should be flexible enough to generate as many sequence reads as desired necessary for the specific research question at hand. In addition, it should be both cheap to acquire and run, easy to operate, have short run time and should involve simple or no library pre-preparation steps.
 Dr Varsha Potdar
Dr Varsha Potdar
Scientist ‘D’ & Influenza Group Leader, ICMR- NIV, NIC, MoHFW
National Influenza Center, Ministry
of Health & Family Welfare
“The ongoing pandemic of COVID-19 has seen the utmost usefulness of whole genome sequencing to track the evolution of the SARS-CoV-2. Next Generation Sequencing platforms that are playing a major role include Illumina, Ion Torrent, and Nano pore. Choice of a particular platform is based on the application, throughput, quality and cost effectiveness. The ICMR-National Institute of Virology, Pune has multiple platforms in place for the SARS-CoV-2 genome sequencing since the start of the pandemic in February 2020. From our experience it has been observed that differing sequencing protocols and use of pipelines for the genome reconstruction and variant analysis may result in discrepancies. Genome quality in terms of per base sequence quality and overall coverage is vital for accuracy of inferences made during variant calling. As an end user, to facilitate use and portability of different platforms it would be appropriate to have uniform sequencing protocols and post sequencing analysis pipelines. This would result in cost effective solutions in terms of supply of sequencing reagents uninterruptedly facilitating desired quantum of genome sequencing efforts which is the need of the hour, in a country like India.”
The incorporation of Artificial Intelligence (AI) has increased the utility of genomic data in clinical practices and thus creates a new frontier in the advancement of genomic data analysis. In May 2020, Nvidia Corporation developed new AI tools, genomic sequencing software, and speech recognition technologies to aid the COVID-19 research efforts. Thus, AI accelerates the variant annotation and prioritization in the clinical diagnosis, which drives the market.
 G R Chandak
G R Chandak
Chief Scientist
CSIR-Centre for Cellular and Molecular Biology
 Karthik Bharadwaj
Karthik Bharadwaj
Scientist, Principal Clinical Geneticist,
CSIR-Centre for Cellular and Molecular Biology
“The next generation sequencing or NGS the popularly used acronym; is indeed next generation, as you can see the two of us! I am reminded of an incident during postdoc days in early nineties, when one of the co-postdocs from Malaysia said while discussing about PCR and Sanger Sequencing that he will design thousands of primers and actually sequence the genome of rare species. Obviously, he was ridiculed to the extent that we awarded him a Nobel Prize!
Looking back, I feel indeed a new generation has heralded within just three decades; be it, population genomics, genetic disease risk, or understanding basic characteristics or evolutionary perspectives or functional aspects. The major impact has been in the health sector, especially those involved in providing medical services. It is a matter of time that it gets organised into hub & spoke model such that physicians prescribe the tests, the data comes from central large sequencing centres, bioinformatic units and labs in hospitals analyse the data and generate reports in mutual consultation with physicians. The hospitals would invest in low throughput sequencers for obtaining results of immediate relevance such as pharmacogenetics, microbial genomics or small panel sequencing for various applications. Probably, sequencers with medium throughput will disappear as the cost of sequencing keeps coming down. Sadly, no middle path for NGS users.”
With a significant decrease in cost coupled with the availability of funding, several start-ups have entered the market. For instance, in January 2020, Omniome, a startup firm from the US, raised USD 60.0 million in Series C financing to speed up the late-stage product development and product launch of its genetic sequencing platform, which consequently fuels market progression.
Since the completion of the Human Genome Project, the DNA sequencing cost has declined precipitously. The cost of decoding one person’s genome was around USD 50,000 a decade ago and currently has reached nearly USD 600 in the largest genomic centers. Recently, in February 2020, BGI claimed to reduce the cost to sequence the genome to USD 100. This is expected to expand the use of this technology for prenatal tests, cancer screening, and research studies.
Globally, there has been commencement of several big genome projects that will churn out data of immense potential. The most ambitious one being the internationally collaborated Earth BioGenome Project (EBP), a moonshot for biology that aims to sequence, catalog, and characterize the genomes of all of Earth’s eukaryotic biodiversity over a period of 10 years. Scientists realized that out of ~2.3 million species that are actually known, only fewer than 15,000, mostly microbes, have been completely or partially sequenced. Thus, understanding the overall life relationship across Mother Earth would not be feasible without accommodating all of them.
The idea itself emerged in the year 2015 but the strategic blueprint of sequencing at species-level not genus was formulated at a global meeting in 2017. No surprise that Asia’s top scientific countries like India and China are also part of this global initiative. Additionally, these two nations have also initiated several other big indigenous NGS projects to explore local biome. One such project in China is aimed to collect DNA samples of 10 percent (70 million) of its male population. The idea is to catalogue the polymorphism associated with Short Tandem Repeats on the Y-chromosome (Y-STRs) to be useful for forensic studies. With acquisition of huge sequencing data (~200 petabytes in case of EBP) at unprecedented rate, the shortage of storage would not seem much problematic as commercial partners like Amazon Web Service, Google, and others are also backing the efforts. However, shortage of skilled personnel for analysis of data along with legal and ethical issues of data ownership may hinder its value-based utilization.
In the field of disease-associated genetic screening, the NGS is highly useful in identifying the monogenic diseases with locus heterogeneity, such as hereditary cancers, mitochondrial diseases, etc. Going by the example of the recent global spread of SARS-CoV-2 virus that led to COVID-19 pandemic, the segment of pathogen diagnostics; understanding susceptibility to and resistivity against the infection; and genetic screening application is also expected to grow very fast in coming years. However, with such advent in disease genomics, in order to provide a customized treatment, the promising concept of genome-based precision medicine will see it future course of realization.