DNA Sequencers
Sequencing business poised for a change

It is time to look at the DNA sequencing landscape with fresh eyes.
For DNA sequencing, this is the year of the big shake-up. From basic biology to virology to human evolution, sequencing is critical in all of these domains, and its significance is only increasing. Biologists are constantly coming up with new techniques to utilize genomics to analyze single cells, and clinicians are itching to harness it for early detection of cancer and other disorders. But for years, most sequencing has relied on machines from a single company, Illumina.
At a conference, Advances in Genome Biology and Technology in Orlando, Florida, a startup business, Ultima Genomics, recently claimed that it could provide human genomes for USD 100 per pop, which is one-fifth the going charge, using novel twists on old technology. At the same conference, several other companies also made promises of quicker, more affordable sequencing. Important patents defending Illumina’s sequencing technology will expire this year, opening the door for additional competition. One such competitor is MGI, a Chinese firm that stated in June 2022 that it will start selling its instruments in the United States this year. The next sequencing revolution could be just around the corner for business.
The majority of sequencing businesses, including Illumina rely on sequencing by synthesis. The DNA that needs to be decoded is split into single strands, which are then often cut into little bits and put on a surface, frequently a tiny bead, in a device known as a flow cell. Each fragment of a single strand acts as a template to direct the creation of a strand with complementary bases that are given one at a time to bead channels. Each additional base has been changed to glow, so a camera can record where it attaches and, in turn, identify the base to which it corresponds on the original strand. Up until the new DNA strand is finished, the procedures are repeated.
Ultima streamlined the process by spraying the DNA-laden beads by the billions onto round silicon wafers, the size of dessert plates. Nozzles above each wafer gently squirt out bases and other reagents, which spread thinly and evenly across the wafer as it rotates, reducing the amount of these expensive materials needed. Instead of moving back and forth across under the camera, the disk moves in a spiral, akin to how a compact disk is played, which speeds up imaging. It is clever engineering that avoids a lot of complex plumbing. A neural network program rapidly turns imaging data into a sequence.
Additionally, the sequencing chemistry is different. Few bases have fluorescent tags, which lowers costs. Additionally, the bases are missing the customary stop signal, ensuring that no excess bases attach. The developing chain can occasionally add numerous bases at once without these terminators, hastening the process. Although many of these advancements are employed elsewhere, they seem to have worked quite well together in this context.
The technology’s promise was proved in four preprints published in late May on bioRxiv by Gilad Almogy, CEO, Ultima, and his coworkers. In one, they utilized their machine to sequence more than 224 previously sequenced human genomes with colleagues from the Broad Institute of MIT and Harvard, and the results were comparable to the earlier research. The three additional studies demonstrated that the method can assess mutational effects, the effects of epigenetics – chemical alterations of DNA that affect gene activity – and the repertoire of expressed genes in a single cell.
In another preprint, Joshua Levin and his Broad Institute colleagues tested the ability of the Ultima technology to identify active genes in single blood cells as indicated by the genes’ RNA transcripts. The team found Ultima’s machine identified those genes about as well as Illumina’s did. And, it is a game changer due to the lower cost.
Fluorescence in situ hybridization
 Pradeep Kasthuri
Pradeep Kasthuri
Business Development Manager – Flow Cytometry,
Sysmex India Pvt. Ltd.
Cytogenetics entered the molecular era with the introduction of in situ hybridization, a procedure that allows researchers to locate the positions of specific DNA sequences on chromosomes. Since the first in situ hybridization experiment in 1969 (Gall & Pardue, 1969), many variations of the procedure have been developed, and its sensitivity has increased enormously. Today, most in situ hybridization procedures use fluorescent probes to detect DNA sequences, and the process is commonly referred to as FISH (fluorescence in situ hybridization). A variety of FISH procedures are available to cytogeneticists, who use them to diagnose many types of chromosomal abnormalities in patients. The success of FISH, and all other methods of in situ hybridization, depends on the remarkable stability of the DNA double helix.
It is well known that most hematological disorders are caused by genomic changes, such as point mutations, chromosomal rearrangements, copy number variations (CNVs), or a combination of these. Technological advances have enabled us to uncover disease-driving mutations and translate these findings to actionable targets.
OGT, a Sysmex Group company, is a leading global provider of clinical and diagnostic genomic solutions that are created for scientists by scientists – including CytoCell®, CytoSure®, and SureSeq™ ranges of FISH, microarray, and NGS products. The company is dedicated to creating products that enable researchers and clinical decision makers to reach the right care decisions for each patient, every time. OGT strives to unlock the future of genetic clinical care with a commitment to working in partnership with its customers – not only by sharing its expertise of 25 years at the forefront of genetic endeavor but also by working closely with scientists to understand their unique challenges, and to customise its approach to meet their exact needs. Dedicated to improving clinical care, OGT believes that through partnership – together – we will achieve more.
OGT, the molecular genetics company, has further expanded its extensive catalogue of FISH products with new CytoCell® probes. The range now includes ETV6 Proximal Probe Green, ETV6 Distal Probe Red, MPO Probe Red, TP53 Probe Green, NUP214 Probe Red, DEK Probe Green, TAS2R1 (5p15.31) Probe Green, Chromosome 9 Satellite III Probe Aqua, and Chromosome 15 Alpha Satellite Probe Red. These cost-effective and quality-assured probes produce specific, high-intensity signals with excellent contrast and minimal background for the utmost confidence in results. Optimized for use on common sample types, the probes are also pre-mixed in hybridization buffer, increasing the ease-of-use and reducing errors in the laboratory.
Computational biologist Lior Pachter, who works at the California Institute of Technology, is skeptical of the new technology. Doctoral student A. Sina Booeshaghi and he examined one of the most active genes in Levin’s team’s blood cells, which may serve as a cancer biomarker and also generate a protein that some athletes use illegally to boost their performance. Sometimes the active gene was missed by the Ultima technology. According to Pachter, both the performance and the mistake rate were extremely high. Pachter and others disagree with the claimed price of USD 100. That sum only accounts for chemicals; it does not include labor costs, pre- and post-sequencing procedures, or the machine’s initial investment, the cost of which has not been disclosed. Even if the USD 100 figure is real, it may not be unique – other companies are also promising USD 100 per human genome.
MGI, a subsidiary of Chinese sequencing giant BGI. MGI’s technology is similar to Illumina’s, but it increases accuracy by adding all four bases at once as it sequences DNA. To track which bases are incorporated, it uses antibodies, which are brighter and less expensive than fluorescent dyes. Illumina, too, is promising lower costs, and at the meeting it introduced new chemistries to increase accuracy and flexibility.
Other companies with new flow cells and chemistries can economically sequence small numbers of genomes. Recently, Molly He, CEO, Element Biosciences reported that the company is now shipping bench-top sequencers that can sequence three human genomes at a time, at a cost of USD 560 each. Another company, Singular Genomics, also promises bench-top technology that does not require high throughput for cost savings.
These machines, like those from Illumina, MGI, and Ultima, all decipher short fragments of DNA. But for the past seven years, two companies, Pacific Biosciences and Oxford Nanopore Technologies, have worked on sequencing long reads, thousands of bases long, which leave fewer partial sequences to piece together into a full genome. The technologies can sequence the native DNA molecule, in all its glory. They have struggled with low accuracy and high cost, but they are on their way to becoming practical tools.
Do not write off the dominant sequencing company Illumina just yet. To maintain a strong position in the market, its scientists likely kept a few cards in their back pockets. Nevertheless, Illumina will encounter unheard-of competition; it is necessary to adopt a fresh perspective on the DNA sequencing market.
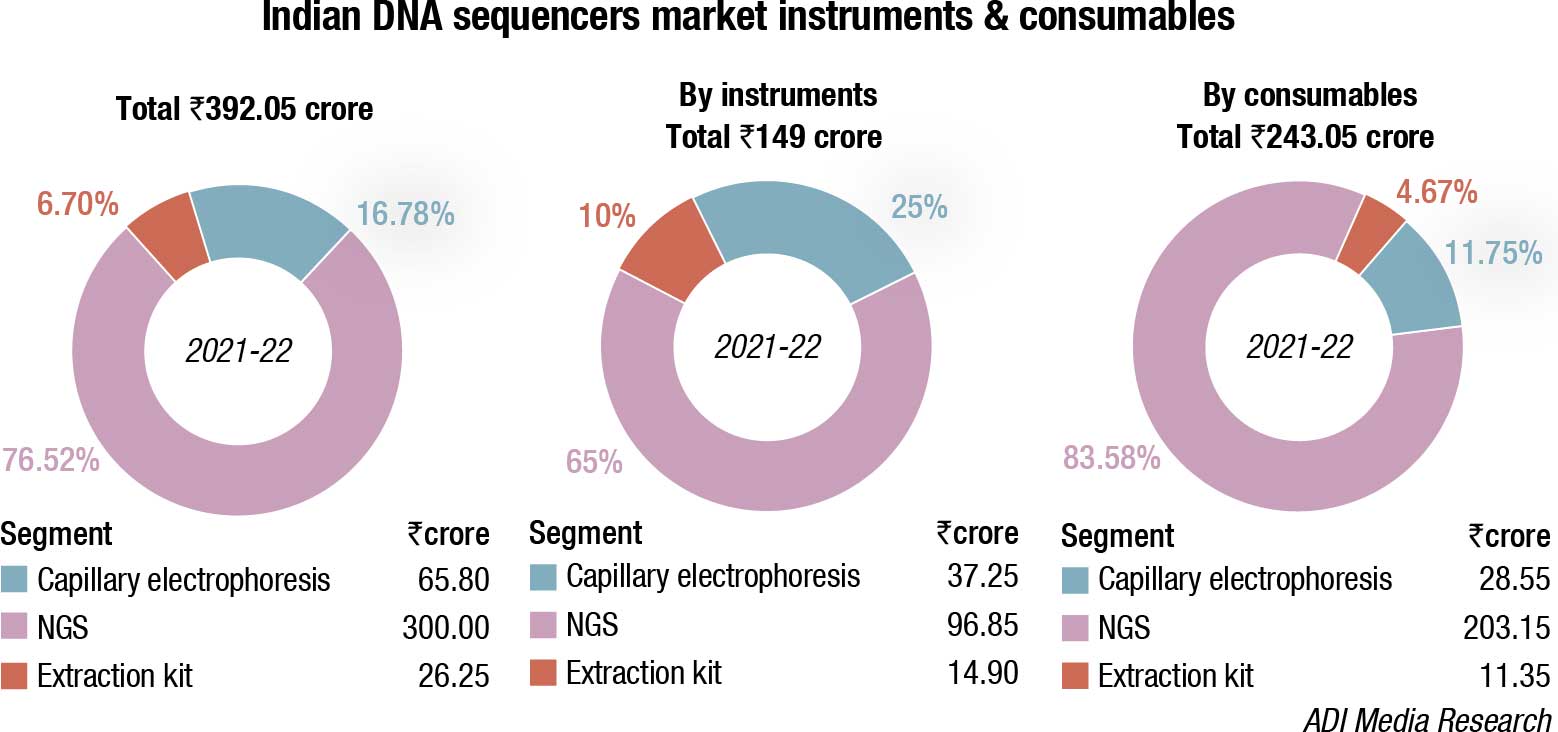
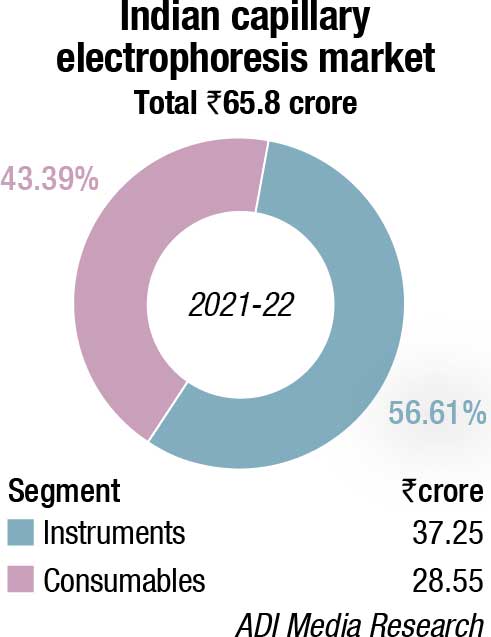
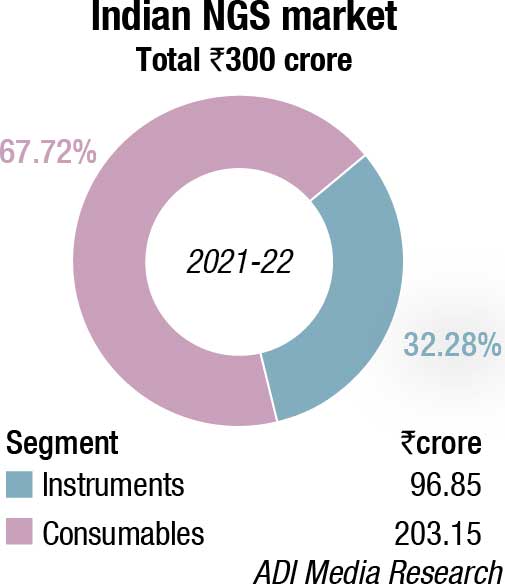
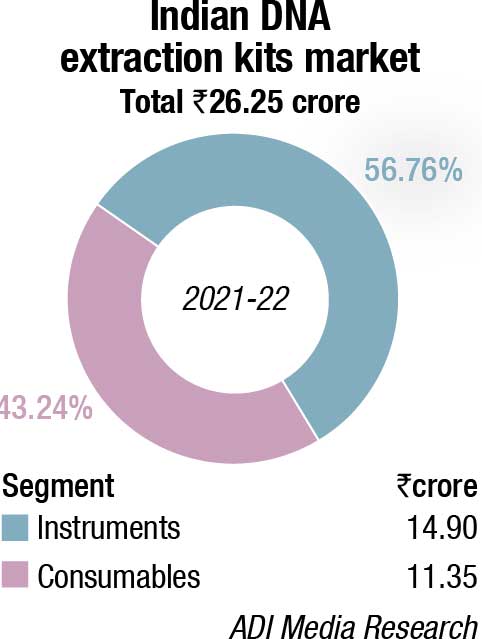
The DNA sequencers market made a comeback in 2021-22, after having seen a sharp drop of 31.5 percent in 2020-21. This may be attributed to the increasing relevance of NGS that increased its segment contribution from 67 percent in FY21 to 77 percent in FY22. The percent share of both CE and extraction kits declined from 18 percent to 16.7 percent and 14.5 percent to 6.7 percent, respectively, in FY22 over FY21. Instruments constitute 38 percent; the balance being contributed by consumables.
This scenario may change in FY23. In the two years of Covid, the laboratories across the county were equipped to handle ID surveillance, also propped up by funding received from Bill & Melinda Gates-UNICEF and WHO. These labs are no longer able to use their equipment to the extent Covid demanded, nor are they skilled to graduate to NGS.
|
Indian DNA sequencers market |
|||
|
Leading players – 2021 |
|||
| Instruments | Consumables | ||
| Capillary Electrophoresis | Tier I | Thermo Fisher (brand Applied Biosystems) |
Thermo Fisher |
| Tier II | NimaGen, Qiagen, GE, Millipore, Chinese brands, and home brewed labs | ||
| NGS | Tier I | Illumina (Premas Life Sciences) | |
| Tier II | Thermo Fisher, Pacific Biosciences (Spinco), and Agilent | ||
| DNA Extraction Kit | Tier I | Qiagen | |
| Tier II | Thermo Fisher and Promega | ||
| ADI Media Research | |||
Demand is there, since India is reporting increasing healthcare-associated infections (HAIs). Often caused by multidrug-resistant organisms on account of indiscriminate use of antibiotics, these are being referred to as a pandemic. Surveillance for HAIs is important to measure their burden, identify high-risk populations and procedures, and guide efforts to reduce HAI incidence. HAI surveillance is a core component of infection prevention and control programs.
Virus pandemics have happened, are happening, and will happen again. In recent decades, the rate of zoonotic viral spillover into humans has accelerated.
Genetic detection, identification, and characterization of infectious agents using NGS has been proven to be a powerful tool. The issue is the Center and Indian states need to create/allocate funds for this, and also train the lab staff. Private philanthropy can be expected to play a crucial role in bridging the gap.Private domestic giving has grown at a moderate pace of 8–10 percent over the last couple of years, CSR being an important component. However, it brings with it compromise on data privacy, and that will need to be addressed, for that has a direct impact on areas as medical tourism and foreign direct investment, among others.
Prime Minister Narendra Modi had recently announced that India’s genomic sequencing network would be extended to neighboring countries through Indian SARS-CoV-2 Genomics Consortium (INSACOG). India’s genome sequencing efficiency and capacity has improved over the last two years, and it now ranks tenth in genome sequencing.
INSACOG initially comprised ten government laboratories. Over a year, it has expanded to 38 centers, including private laboratories. Having said this, the consortium has been slow to sequence significant numbers of samples. It has also not been able to effectively deploy sequencing information to prepare for and prevent future possible waves of infection. This is partly because of INSACOG’s own inability to establish clear systems for constituent laboratories and partly because of political apathy to the information provided by the consortium.
India will need to focus on genomic sequencing, and use of scientific evidence for decision making is not a choice but an absolute essential.
India formed the Indian SARS-CoV-2 Consortium on Genomics (INSACOG), a consortium of 10 genome sequencing labs, in December 2020 to pump up efforts for genome sequencing. The consortium was jointly initiated by the Union Ministry of Health and Department of Biotechnology (DBT) with Council for Scientific & Industrial Research (CSIR) and Indian Council of Medical Research (ICMR). Later, 33 more research laboratories were included in the network to enhance sequencing efforts in the country. These 43 laboratories operate on a hub-and-spoke model, where the 10 INSACOG Genome Sequencing Laboratories (IGSLs) guide new laboratories and act as hubs.
In an order dated February 22, 2022, the Department of Biotechnology (DBT) approved the inclusion of six private laboratories in the consortium. These include Strand Life Sciences, Genotypic Technologies, Medgenome, and Eurofins Genomics India – all based in Bengaluru – along with the Delhi-based CARINGdx and Neuberg Supratech Reference Laboratories, Ahmedabad.
In July 2022, INSACOG announced that it is looking to expand its network of labs to neighboring countries. Indian government officials have chalked out a roadmap on the kind of support India can extend to neighboring nations in training, capacity building, and building facilities as well as carrying out research in India with samples given by these countries.
Dr Rajesh S. Gokhale, Secretary, Department of Biotechnology (DBT) at the Union Ministry of Science & Technology, said, “A meeting was held between DBT and MEA. We have set up a roadmap. This roadmap will allow us to work with all the neighboring nations in the best possible way they are comfortable with and what all India can provide in terms of fulfilling their requirement. The discussions were held at all levels and it will vary with all the places. For instance, in the past, we have helped Sri Lanka to do genome sequencing because they have some institutions to do this activity. However, Bangladesh might require a proper set-up and computational power as well. Some countries may ask for some aspects of things. Maybe if they are interested to come and learn in India, then we can bring them to our labs.”
“DNA sequencing refers to the general laboratory technique for determining the exact sequence of nucleotides (As, Ts, Cs, and Gs), or bases in a DNA molecule. In Sanger, sequencing is based on fluorescent chain terminator nucleotides that mark the ends of the fragments and allow the sequence to be determined. The target DNA is copied many times, making fragments of different lengths. Sanger sequencing gives high-quality sequence for relatively long stretches of DNA (up to about 900 base pairs). Next-generation sequencing (NGS) techniques are new, large-scale approaches that increase the speed and reduce the cost of DNA sequencing. There are a variety of NGS techniques that use different technologies. However, most share a common set of features that distinguish them from Sanger sequencing. Highly parallel and micro scale, many sequencing reactions take place at the same time in tiny chips; reads typically range from 50 to 700 nucleotides in length; and it is fast and cost-effective. Future belongs to NGS. The Covid-19 pandemic has brought qPCR and NGS to so many labs where normal PCR was also unavailable. Precision medicine, cancer diagnostics, surveillance in pandemic will be a few key areas. With technology advancement and lower per GB cost, it will be widely used in clinical domain. Dr Varsha Potdar, Scientist ‘D’ and Influenza Group Leader, ICMR – National Institute of Virology, National Influenza Center, Ministry of Health & Family Welfare.
The consortium in its attempt to answer questions related to host immune response, long-term effects in immunity of Covid-19 infected individuals, is working toward establishing a hospital network across the country. This arm of the consortium aims to study clinical correlations in mild versus severe cases of Covid-19, longitudinal study to understand long-term post-Covid complications and change in immunity. Additionally, the consortium is also looking to expand to sewage surveillance as a quick and early detection tool of assessing the spread of variants in a hotspot locality of increased positivity. However, the consortium was slow to sequence significant numbers of samples. It has also not been able to effectively deploy sequencing information to prepare for and prevent future waves of infection. This is partly because of INSACOG’s own inability to establish clear systems for constituent laboratories and partly because of political apathy to the information provided by the consortium. The INSACOG project has come to exemplify the government’s approach to scientific counsel in tackling the pandemic – to ignore it.
According to data uploaded on the INSACOG portal, from the time INSACOG was formed till the end of January 2022, Maharashtra sequenced 24,704 samples, while states like Bihar and Jharkhand sequenced only 594 and 783 respectively. So, more than 25 percent of sequencing data in India was from Maharashtra alone. The resulting national-level sequencing data and prevalence of new variants mostly reflects what is going on in the states that are sequencing much more than the rest of the country.
The INSACOG data portal has been up since early 2021, soon after the consortium was formed. But data shared through the portal remains limited and is not updated in real time. Since February 2022, this portal is no longer being updated and INSACOG data is hosted on a website run by the Indian Biological Data Center, a website that has restricted access and has very limited data available for the public.
“Analysis of DNA polymorphism in specific genes by targeted amplicon sequencing (TAS) has come up as a highly accurate analytical tool for testing identity, paternity, fidelity, disease diagnosis, breeding, and predisposition to genetic disorders.
The available TAS platforms include IonTorrent’s Ampliseq and Illumina’s target capture MiSeq for medium sample throughput medical diagnostics and plant breeding applications. Both the platforms were utilized heavily for complete viral genome sequencing of SARS-CoV-2 genome during Covid-19 pandemic. AgriSeq genotyping platform, specially designed for plant genetics and breeding applications, is validated for multiplexed PCR amplification of 100–5000 SNP markers in a single tube. Target-captured high-sample throughput deep-coverage amplicon sequencing has also been optimized on NovSeq and NextSeq NGS platforms. Bar coding allows pooling of samples from different species or individuals in a single sequencing run. TAS maintains a high concordance, as compared to chip array-based genotyping, due to deep sequence coverage. In-built data analysis suites allow easy visualization and summarization of the results. Nagendra Singh, National Professor, ICAR-NIPB, New Delhi.
Before the pandemic, India’s genetic sequencing efforts were limited to particular laboratories and conducted sporadically. TB is one pathogen for which there was a nationwide effort to sequence. There were many samples collected at a national level but very few laboratories in the country that were associated with the actual sequencing effort.
But regardless of whether India consistently increases its sequencing capacity or meets the aspirational five percent target, INSACOG will have to streamline its strategy and implement it uniformly across the country and work in order to inform timely public health intervention.
The majority of scientists who are working in various INSACOG facilities report that the INSACOG-mandated sequencing strategy is constantly changing and that they are using different versions of it. Additionally, this plan must offer high-quality data that is related to clinical information. In order for administrators and public-health officials to use the data, it must be uploaded and analyzed quickly. All of this also depends on how well the government cooperates with the scientific community, giving them the tools they need to do their jobs and responding quickly when they are alerted to alarming new mutations.
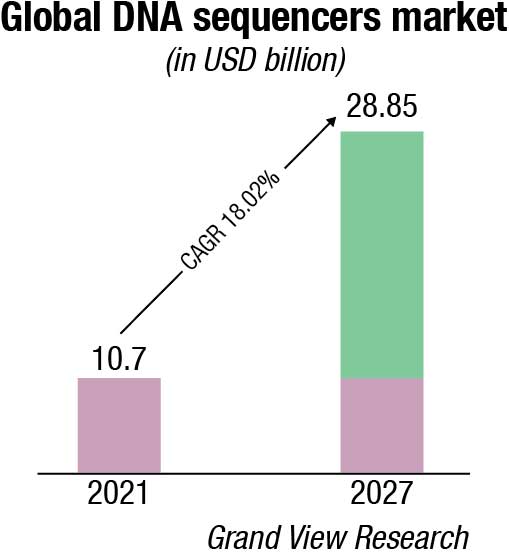
The global market for DNA sequencers is expected to grow from USD 10.7 billion in 2021 to USD 28.85 billion by 2027, at a compound annual growth rate (CAGR) of 18.02 percent for the period 2021 to 2026. Over the years, various sequencing methods, such as Sanger sequencing, next-generation sequencing, and third-generation sequencing have been widely used to determine genome sequencing in less time and in a cost-effective manner. One of the key factors driving global market growth is declining costs of genome sequencing and rapid advancements in next-generation sequencing platforms. In addition, DNA sequencing is widely used in applications, such as biomarker discovery, reproductive health, agrigenomics, forensics, and HLA typing, among others.
One of the most prominent factors driving the demand for DNA sequencing market is the increasing prevalence of genetic disorders. As per the Ministry of Health and Family Welfare, India (2022) there are as many as 7000 rare diseases across the globe, out of which 72 percent are of genetic origins. The data from the World Economic Forum (February 2020) mentioned that nearly 10 percent of the global population (475 million) people were affected by a rare condition. Furthermore, as per the Department of Health & Social Care, United Kingdom (2022), rare diseases though quite uncommon individually, are collectively quite common. One in 17 people are affected by a rare condition across the globe. In the UK alone, rare diseases affect over 3.5 million people. Rare diseases are often difficult to identify, and either remain undiagnosed or result in misdiagnosis. DNA sequencing like whole genome sequencing offer advantages, such as detection of multiple variant types in a single assay, thereby providing improved diagnostic field and operation efficiency. Considering such benefits of DNA sequencing, some countries are extensively focusing on genome programs in order to provide better healthcare facilities to patients. Thus, the market for DNA sequencing is expected to witness remarkable market growth in coming years with the increasing prevalence of rare genetic disorders collectively and the increasing emphasis on identifying the genetic mutations.
Furthermore, another key contributor in the growing popularity of DNA sequencing is the increasing prominence of precision medicine in cancer care. The concept of precision medicine or individualizing the treatment plan according to the biological behavior of the tumor is considered a new approach in cancer management. The clinical applications of precision medicine are diverse, encompassing screening, diagnosis, prognosis, prediction of treatment response and resistance, early detection of recurrence/metastasis, and biologic cancer stratification. Precision medicine in cancer care is aimed at bringing about therapeutic effects in subsets of patients displaying specific molecular/predictive biomarkers. The customization of treatment specific to the patient’s genetic makeup has gained widespread acceptance and attention from physicians. DNA sequencing has been a key component in driving the concept of precision medicine. Advanced sequencing techniques, such as next-generation sequencing, help map the oncogenes and also in tumor analysis and help develop specific therapeutic agents. Moreover, advantages associated with the technologies, such as the simultaneous analysis of a broad spectrum of genetic alterations, including copy number variations, mutations, fusions in multiple genes, and translocations. This resulted in more personalized drugs being approved by regulatory bodies across the globe. For instance, as per the US Food and Drug Administration, out of the total drugs approved in the country in 2020, 39 percent were categorized as personalized medicine. Therefore, the growing prominence of precision medicine and its advantages in disease management are expected to contribute to the growth of DNA sequencing market in years to come. Additionally, various market players are heavily investing in developing enhanced, cost-effective consumables and portable sequencing technologies. This is providing a major thrust to the global market.
However, technology-related challenges, such as sequence alignment, problems with mapping multiple reads, and high cost of technology may prove to be restraining factors to the DNA sequencing market growth.
Unlike majority of the markets, the DNA sequencing market witnessed a positive trend during the Covid-19 pandemic. There was an exponential increase in the demand for DNA sequencing products and services across the globe due to the exigent need for sequencing the SARS COV-2 virus genome. Even though other domains, where DNA sequencing is employed, were affected by the lockdown restrictions, the dire need for sequencing the genome of the novel strain of the SARS-CoV-2 virus and the subsequent development of vaccines kept the momentum of the DNA sequencing market. With the return of normalcy across the globe, the future outlook for the DNA sequencing market looks positive in coming years.
In the technology segment of the DNA sequencing market, the long-read sequencing category is expected to show tremendous potential in revenue growth over the next 5 years. This can be ascribed to the advantages associated with long-read sequencing over other DNA sequencing technologies. Long-read sequencing is the emerging technology in DNA sequencing and offers various advantages over NGS and Sanger sequencing. Short-read sequencing, though cost-effective and accurate, also becomes challenging in terms of reconstructing and counting the fragments. Such limitations are overcome by long-read sequencing as the technology allows for reading longer stretches of DNA, subsequently helping de novo assembly, mapping certainty, transcript isoform identification, and detection of structural variants. Two most prominent long-read sequencing technologies are SMRT® sequencing technology by Pacific Biosystems and Oxford Nanopore Technologies. Thus, considering the advantages associated with long-read sequencing technology, this category is estimated to perform well in coming years.
Next-generation sequencing is projected to dominate the technology segment of DNA sequencing market revenue. The application of DNA sequencing in developing diagnostic tests and vaccines for viral diseases, such as the SARS CoV-2 has augmented the demand for DNA sequencing exponentially. Thus, high incidence of novel viral diseases and ongoing research and development activity in DNA sequencing are major reason for the high revenue share of next-generation sequencing.
North America is expected to dominate the DNA sequencing market. Among all the regions, North America is estimated to amass the largest revenue share in the global DNA sequencing market. This can be ascribed to the high prevalence of cancers, and a supportive regulatory environment, among other factors, in the region. Furthermore, high disposable income, sophisticated healthcare infrastructure, and increased awareness regarding new sequencing methods and programs are also expected to aid in the DNA sequencing market growth in this region.
One of the key supporting factors for the growth of the North America DNA sequencing market is the increasing prevalence of cancers in the United States. As per the figures mentioned by the American Cancer Society, in 2021, it was estimated that 1.9 million new cancer cases would have been diagnosed in the United States. The data provided by the Centers for Disease Control and Prevention (2021), 1,708,921 new cases of cancers were reported. For instance, the American Cancer Society estimates that in 2022, approximately 287,850 new cases of invasive breast cancer are expected to be diagnosed in women in the United States. Therefore, the increasing incidence of cancers like breast cancer in the country is expected to further drive the demand for DNA sequencing, such as next-generation sequencing, for tumor analysis and identification of prognostic biomarkers, thereby contributing in the growth of the United States DNA sequencing market as well as the overall growth of the North America DNA sequencing market.
Furthermore, the growing emphasis on genome sequencing like human genome project and also establishing DNA sequences of other infectious microorganisms. For instance, in April 2021, Biden administration announced USD 1.7-billion investment in managing the spread of Covid-19 variants in the country. The funds were aimed at helping the CDC to improve their sequencing abilities to identify variants of the SARS-CoV-2 virus. There is also a key focus on developing a national bioinformatics infrastructure in the country and this investment is thought to support bioinformatics throughout the US public health system, creating a unified system for sharing and analyzing sequence data in a way that protects privacy but allows more informed decision-making.
Some key companies operating in the global DNA sequencing market include 23andMe, Oxford Nanopore Technologies Ltd., Precise.ly, 54Gene, Guardant Health, Allogene Therapeutics, Pacific Biosciences, CardioDx, Helix, Human Longevity, Foundation Medicine, Freenome, Veritas Genetics, Agilent Technologies, Inc., Thermo Fisher Scientific, Inc., Illumina, Inc., Pacific Biosciences, Inc., Qiagen, F. Hoffmann-La Roche Ltd., Macrogen, Inc., Perkin Elmer, Inc., BGI, Bio-Rad Laboratories, Inc., Myriad Genetics, PierianDx, Eurofins Scientific, and Intrexon Bioinformatics Germany GmbH.
For most DNA sequencing companies, the future lies in developing new applications. One of the discoveries that have paved the way is the presence of small DNA fragments in blood. First used for non-invasive pre-natal testing of fetal DNA, companies have now moved on to cancer diagnostics. Illumina now sees itself as an oncology diagnostics developer. The company does not focus just on the sequencer; it has a strong focus on the end-to-end process. The aim is to make sequencing-based diagnostics easy enough to be carried out by non-specialists. Ion Torrent is also developing this approach.
Cancer diagnostics is not the only application for NGS, of course. If there is any silver lining around Covid-19, it has shown what sequencing can do. Traditionally, companies have had less of a focus on infectious diseases. But, the pandemic has changed that situation dramatically.
Illumina developed kits for Covid-19 surveillance, creating a reference genome, against which other viral genomes could be compared. Ion Torrent has also moved into infectious diseases surveillance over the last two years. Oxford Nanopore developed the LamPORE assay, a cheap and highly scalable Covid-19 diagnostic test, which detects the virus in 90 minutes.
The progress of DNA sequencing over the past few decades has been tremendous. But there is undoubtedly much more to come as we continue to learn how to read the book of life. In the words of Sanger’s co-Laureate Walter Gilbert, “I think the future of science will continue to astound us.”