Microbiology Instruments and Reagents
mNGS is transforming the landscape of microbiology laboratories
mNGS is transforming the landscape of clinical microbiology laboratories, and to ensure that it is properly utilized in clinical diagnosis, both physicians and microbiologists should have a thorough understanding of the power and limitations of this method.
In the last 50 years, several diagnostic approaches have been introduced in medical microbiology, including amplification-based (PCR), MALDI-TOF, DNA-microarray-based hybridization technology, T2 magnetic resonance, and next-generation sequencing. However, none of these methods could fully replace standard techniques (microscopy, culture, and serology).
Molecular biology revolutionized the diagnosis of infectious diseases, especially for detecting viruses and identifying bacteria, involved in sexually transmitted infections, gastrointestinal infections, and tuberculosis. Yet, today’s clinical microbiology laboratories have not changed dramatically since the early 2000s. This is mainly due to the advantages of traditional standard techniques. Firstly, cost-effectiveness and extensive clinical validation and secondly, limitations of newer methods, such as limited spectrum, sensitivity and specificity, the lack of differentiation between living and dead cells, and the importance of phenotypic antimicrobial susceptibility testing. Moreover, one existing challenge in diagnostics remains; a priori knowledge of what to expect from a particular clinical sample or patient. In most cases, a priori knowledge is enough to request the most appropriate test, such as multiplexed panels or specific culture media, but this is not always the case.
Clinical metagenomics next-generation sequencing (mNGS) has the potential to surpass many limitations of current routine diagnostics methods. It can reveal information at different levels, including detecting and characterizing all microorganisms and viruses (DNA and RNA), without a priori knowledge from a single test. High expectations have been built around mNGS; however, this technique is far from widely available.
The need for a novel diagnostic tool to increase the sensitivity of microbial diagnostics is clear. mNGS has the potential to revolutionize clinical microbiology. However, its role as a diagnostic tool has yet to be widely established, which is crucial for successfully implementing the technique. A clear definition of diagnostic algorithms that include mNGS is vital to show clinical utility. Similar to real-time PCR, mNGS will one day become a vital tool in any testing algorithm.
Implementation of mNGS in routine diagnostics
The diagnostic laboratory will need to invest in IT infrastructure, separate sample/library preparation areas and equipment, such as micropipettes, validation processes, and NGS-specialized laboratory personnel. The diagnostic laboratory may opt to (partially) outsource mNGS wet- and dry-lab processing to accredited and commercial service providers to negate infrastructure costs, such as dedicated laboratory space, high-performance e-infrastructure, including networks, software stacks, and large-scale storage resources. However, at the same time, routine diagnostic labs may have existing areas for pre-PCR preparation that could be incorporated into the mNGS workflow. It is important to stress that the e-infrastructure should be designed and maintained in a collaborative effort between the diagnostic laboratory and the IT department.
Processes, such as nucleic acid extraction and library preparation, would be ideally automated. Existing platforms, such as extraction platforms, pipetting robots, and thermal cyclers, currently used for molecular diagnostics, can also be integrated into the mNGS workflow. Several studies have evaluated the performance of available nucleic acid extraction platforms for mNGS and have found variable results. Library preparation can initially be performed manually, if the sample throughput is low. The sequencing approach and platform can be selected based on sample throughput. mNGS requires trained laboratory technicians and bioinformaticians/computational biologists, which can increase costs. User-friendly specialized software and pipelines can be utilized for automated analysis. However, results must be validated and interpreted by a multidisciplinary team of medical microbiologists and clinicians.
Turnaround time
The turnaround time of a diagnostic test is desired to be within a clinically actionable time frame. A conventional diagnostic workflow can take a few hours to 2–7 days from sample collection to identification and antimicrobial susceptibility determination, compared to up to 5 days for mNGS. Several factors affect the turnaround time of mNGS. For example, deeper sequencing by limiting the number of samples per run enables more detailed taxonomic resolution, antimicrobial drug resistance prediction, and phylogenetic analysis at the expense of extended turnaround time. The number of samples per run can impact the cost efficiency of mNGS. Running individual or a few samples might be necessary in the event a rapid diagnosis is required. Low-throughput platforms can negate some of the extra costs, but are hampered by the inclusion of positive and negative controls in each run.
Depending on the clinical situation, the implementation of mNGS in routine diagnostics can be advantageous compared to conventional testing by circumventing some limitations, e.g., in identifying uncultivable microorganisms or slow-growing bacteria. Continuous technical advancements in sequencing technologies, particularly real-time ONT sequencing, could accelerate the clinically actionable results in under 6 hours following sampling, e.g., to identify pathogens based on circulating cell-free DNA from blood. Therefore, depending on the intended clinical use, mNGS could be a favorable choice to perform actionable results within a reasonable time.
The targeted pathogen could impact the selection of the nucleic acid extraction method, sequencing strategy (RNA and/or DNA), target enrichment/host nucleic acid depletion requirement, sequencing depth, reference database design, or data analysis tools. Sequencing only one type of nucleic acid may decrease the overall sensitivity of the method, since some viruses may be missed. Furthermore, the complete recovery of bacterial/fungal/parasitic genomes will be unlikely if using RNA-sequencing. To increase sensitivity, some laboratories may opt to sequence all the nucleic acids present in a sample; however, this may increase the overall cost per sample.
The sensitivity of mNGS is hampered by several factors that are dependent on the specimen composition, material, volume (nucleic acid background/pathogen ratio), collection method, transport, and storage. Sensitivity can also be affected by the efficiency of the nucleic acid extraction (bias toward some species), sequencing method, and bioinformatics pipeline used for analysis. On the other hand, specificity is influenced by contaminating nucleic acids in clinical specimens, reagents, or by the accuracy of taxonomical classification algorithms. NGS-related phenomena, such as index hopping or crosstalk, can also introduce false-positive results, resulting in lower specificity. The ratio between host and microbial DNA/RNA is a major determinant of the proportion of microbial reads obtained after metagenomics sequencing. The unbiased nature of mNGS, particularly shotgun metagenomics, leads to the sequencing of background, as well as pathogen nucleic acids.
Challenges
One of the biggest challenges of implementing mNGS is the validation of the test. As with other laboratory-developed tests, the requirements for validation depend on local and federal regulations. Validation is challenging due to the broad nature of the test and a large number of possible results. In addition, often, no reliable reference method with a similar scope is available. Validation is required for both the wet-bench protocols, including accuracy, analytical sensitivity and specificity, reproducibility, stability, as well as bioinformatics protocols. For the latter, in silico analyses using simulated samples can be performed. During validation of the wet bench, it is crucial to define and use proper external and internal controls, which are essential to bring standardization, and ensure the quality of the generated sequences in clinical settings. Despite the challenges to validate mNGS, examples are available for successful implementation for routine testing, such as pathogen detection in cerebrospinal fluid and detection of RNA and DNA viruses in respiratory samples.
Way forward
As many samples remain culture- or PCR-negative, clinical laboratories could benefit from mNGS. However, cost, turnaround time, variable sensitivity/specificity, validation, and reproducibility remain hurdles to overcome before implementing mNGS in routine diagnostics. A commercial mNGS service provider could be applied to reduce costs before investing in infrastructures, equipment, and NGS-specialized laboratory personnel. The analytical sensitivity can be increased by several host-depletion and microbial-enrichment strategies. Reagent and laboratory contamination should be mitigated by sequencing a negative control and post-sequencing contamination removal to increase specificity. Nevertheless, data must be interpreted and evaluated carefully within a clinical context.
Microbiology instruments and reagents in IVD
 Biplab Kumar Sinha
Biplab Kumar Sinha
General Manager – Sales & Marketing,
Beacon Diagnostics Pvt. Ltd. (A Beacon Group Company)
Clinical microbiology has a prominent role to play in the field of IVD and medicines. It supports the fight against infectious diseases and provides valued inputs on antimicrobial resistance. Routine diagnostic tests include microscopy, culture, biochemical identification, and immunological testing. Traditional methodology for these tests is time consuming, labor intensive, and lacks the specificity or sensitivity due to manual involvements.
On the other hand, higher prevalence of infectious diseases and sexually transmitted diseases demands for increase in microbiology testing, and to cope up with this demand more advance methods are emerging in microbiology that focus on speed, accuracy, and economy. Thus, emerging technologies and robust processes have transcended microbiology testing to the next level in IVD.
Rising healthcare expenditure, growing need to speed up microbiological testing, scarcity of time, need for rapid QA-QC procedures, and growing imposition of regulations that regulate various production processes are factors that have catalyzed this transformation. The uptake of microbiology is further favored by technological advancements, including automated instruments to identify pathogens across laboratories. The trend toward automated instrumentation is aided by the promise of overall cost effectiveness and efficiency. Another driver for market growth is consolidation of laboratories and rising consumer awareness.
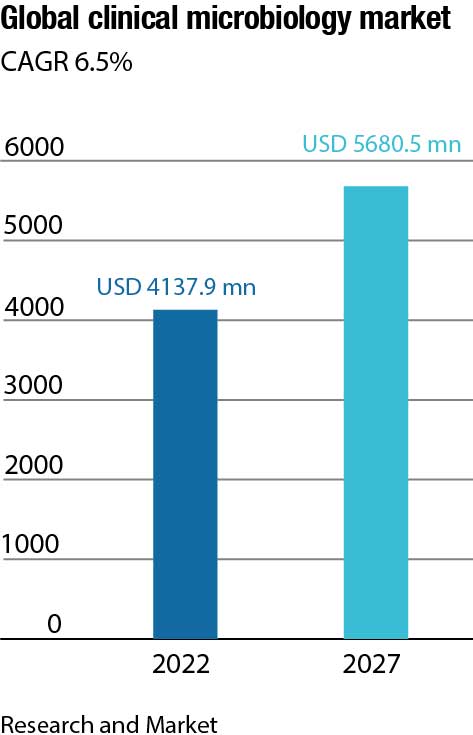
Within the Covid-19 crisis, the global market for clinical microbiology instruments and reagents estimated at USD 4.5 billion in the year 2022, is projected to reach a revised size of USD 5.8 billion by 2026, growing at a CAGR of 6.4 percent.
The evolution pattern of automation in microbiology is already mimicking what clinical chemistry has witnessed in the last 30 years. The challenge for IVD companies will be to provide the solutions with task-specific automation that can be easily integrated with each other via data connections. The companies that can drive automation with new methods, and can create and add value, will gain the edge over other players in this market.
Cost-effective, easy-to-use rapid test for major pathogens, and simple-to-use chromogenic culture media are already making their way ahead in microbiology that aims to make future of microbiology testing precise and more meaningful.
Furthermore, validated microbial community reference standards and external/internal controls are required to warrant quality, reproducibility, and consistency of mNGS workflows. Above all, the intended use of mNGS should be clearly defined and performed on a case-by-case basis as described in the proposed diagnostic algorithm. Additionally, careful consideration is needed to determine the most appropriate clinical approach as each have their own advantages and disadvantages. mNGS can circumvent some of the limitations of conventional testing to obtain a clinically actionable result in a reasonable time frame. Considering the ability of some sequencing platforms to provide same-day results, mNGS can revolutionize routine diagnostics.