MB Stories
Unleashing the power of microbiology

A journey through cutting-edge microbiology innovations, from molecular diagnostics to automation and genomic approaches is reshaping the future.
What microbiology beholds after a decade in the future requires a vision based on the facts and ongoing trends in research and technological advancements. The field of microbiology is moving exceptionally fast, taking full advantage of the new developments in microscopy, computational biology, synthetic biology, lab-on-chip approaches, and single-cell technologies. Advanced microbiology technologies are rapidly changing the ability to diagnose infections, improve patient care, and enhance clinical workflow.
In this ever-evolving landscape of medical science, the realm of microbiology stands at the forefront of pioneering advancements poised to redefine diagnostic methodologies in the next decade. This journey into the future of microbiology unveils groundbreaking developments, each thread intricately woven with precision and innovation. The narrative expands to the corridors of molecular diagnostics, where a paradigm shift from traditional culture-based methods to molecular approaches is witnessed.
As the journey progresses, the role of automation emerges as a pivotal force reshaping laboratory landscapes. Microbiology unleashed stands as a testament to the continued pursuit of excellence in microbiological practices, providing a comprehensive overview of the transformative trends shaping the next decade.
Revolutionizing gut microbiome research – Automated anaerobic systems and future frontiers
 Dr Rahul G Warke
Dr Rahul G Warke
Director – R&D, Microbiology,
HiMedia Laboratories Pvt. Ltd.
We discuss here the significance of the human gut microbiome, particularly the anaerobic bacteria residing in the gastrointestinal tract, and the challenges associated with studying them. The gut microbiota plays a crucial role in health and disease, influencing conditions, such as obesity, inflammatory bowel disease, and metabolic disorders. Traditional methods of culturing anaerobic bacteria have limitations, leading to the emergence of culturomics, a field utilizing various culture conditions and mass spectrometry for identification.
The human gut primarily harbors anaerobic bacteria, making the cultivation and analysis of these microorganisms challenging. The article emphasizes the importance of precise sample processing, collection, and transportation conditions to ensure the viability and diversity of intestinal bacteria. Anaerobic conditions are crucial for cultivating these bacteria, with oxygen levels in the intestine measured below 1 mm Hg.
We envisage automated anaerobic jar systems (AAJS) as a solution to the challenges posed by traditional methods. These systems offer a sophisticated and efficient approach to establishing and maintaining anaerobic conditions for the cultivation of anaerobic microorganisms. The AAJS ensures precision and control over gas mixtures, allowing for consistent and reproducible experimental settings. They also provide high throughput, enabling the handling of multiple samples simultaneously, expediting research.
Advantages of AAJS include their ability to culture difficult-to-grow microbes, time efficiency, and assured results. These systems prove valuable in gut microbiome research, enabling microbial diversity analysis, functional analysis of metabolic activities, drug screening, and disease modeling. The article suggests that AAJS technology will continue to advance, becoming a potent tool not only in gut microbiome research but also in broader applications, such as clinics, deep-ocean and water sediment research, and investigations into various animal gut ecosystems.
Future developments are anticipated to involve improved automation, integration with omics technologies (metagenomics, metatranscriptomics), and the incorporation of artificial intelligence for data analysis and interpretation. The success in anaerobic gut microbial studies is expected to contribute to the growth of the probiotics industry, leading to a steady expansion of the global anaerobic market in the coming years.
Indian market dynamics
The Indian market for microbiology instruments and reagents in 2022 is estimated at ₹650 crore, a 38-percent increase over 2021, the previous year also having seen an increase of 34.3 percent over 2020. The instruments, along with instrument-based reagents in 2022, increased their combined share from 56.4 percent in 2021 to 73.8 percent in 2022, with instruments at ₹80 crore, and the reagents at ₹400 crore. The non-instruments-based reagents declined from ₹212 crore in 2021 to ₹170 crore in 2022.
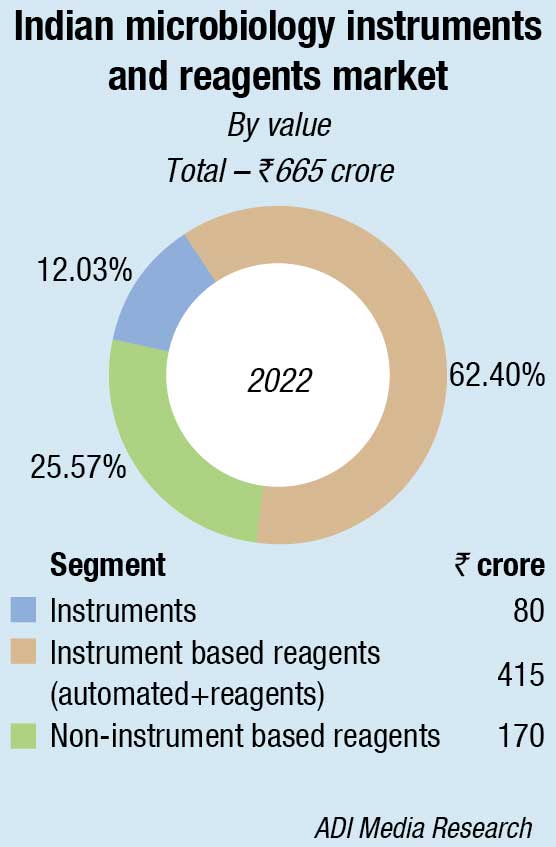
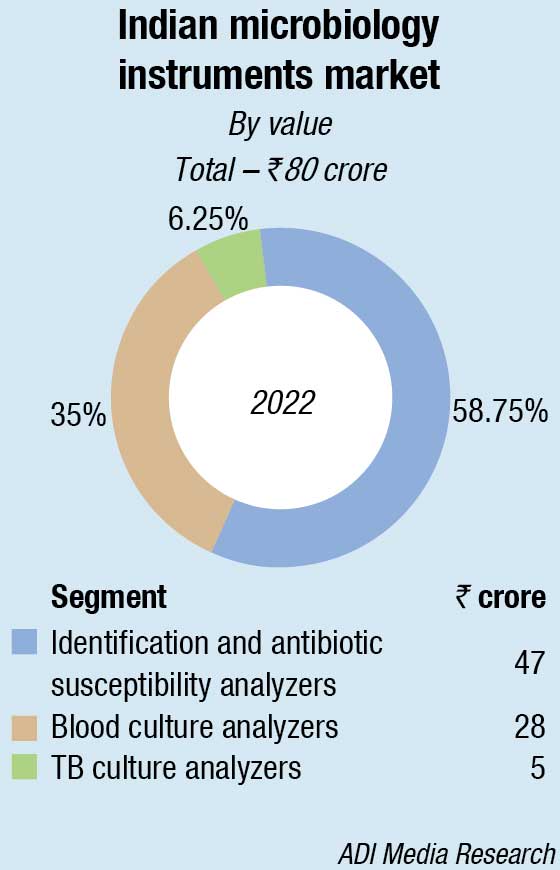
Within the microbiology analyzers segment, in 2022 too as last year, the blood culture analyzers and identification and antibiotic susceptibility analyzers dominated, with TB culture analyzers seeing a steady decline. bioMérieux and BD continue to dominate the instruments and instruments-based reagents segments.
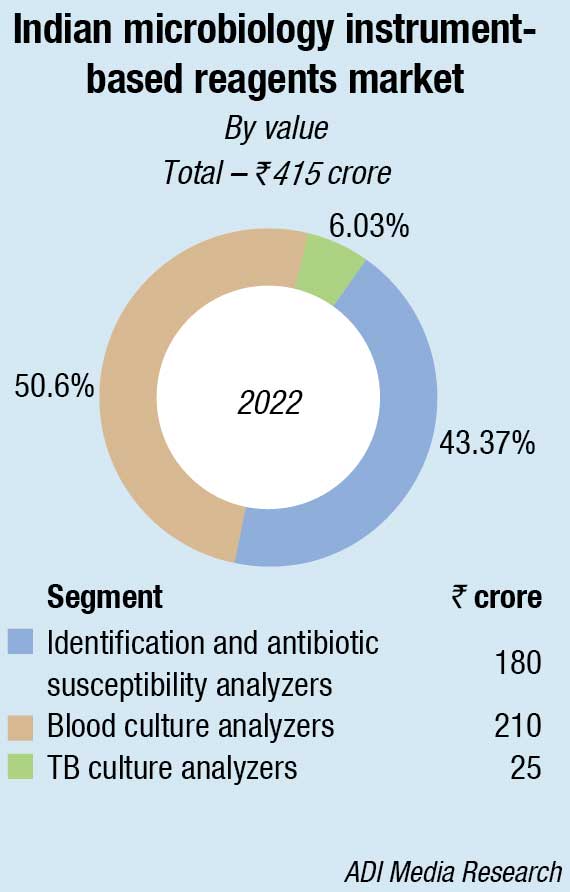
In the non-instruments-based reagents segment, Hi-Media dominates with an estimated 58-percent share. This includes ready-to-use plates, antibiotic disks, MICs, etc. Titan Biotech (Microexpress) and Tulip are the other aggressive players.
With the Indian government having announced its plans to build at least one major hospital in each of India’s 761 districts, 2022 saw large orders from some state governments. And the exercise is on, with some north-eastern states and southern projects in the pipeline.
Infectious diseases (ID) blocks are being set up in the district hospitals, with an overall intent to step up the fight against IDs, especially antimicrobial resistance (AMR). Implementing National Quality Assurance Program (NQAP) and Kayakalp program in public healthcare facilities is hugely contributory to strengthen antimicrobial stewardship (AMS) activities in district and sub-district hospitals in India. These cover aspects such as infection control, standard treatment guidelines (STGs), prescription auditing, essential medicine list (EML), availability of antimicrobials, and incentives for meeting quality standards. Steps are being taken to revise the EML, based on WHO AWaRe classification, incorporating the STGs for common clinical infections from the WHO AWaRe antibiotic book and Indian Council of Medical Research (ICMR) program-mandated requirements for dedicated staff/standards for AMS activities and antimicrobial-specific prescription audits as per WHO AMS tool-kit and ICMR AMS guidelines, identified for strengthening AMS activities. Further, hindrances in executing existing policies are also being addressed, such as shortage of human resources, reluctance to follow STGs, and limited availability of diagnostic microbiology laboratory services.
|
Leading players in the Indian microbiology instruments |
|
| Segment | Leading brands |
| Automated instruments | BioMérieux, BD India and Beckman Coulter; Thermo Fisher and Trivitron |
| Instruments-based reagents | BioMérieux, and BD India; Beckman Coulter and Thermo Fisher |
| Non instruments-based reagents | HiMedia, BD India, Thermo Fisher, Bio-rad, and Microxpress; BioMérieux, Beckman Coulter, Merck Millipore and Titan Biotech |
| ADI Media Research | |
Additionally, growth is fueled by the increasing prevalence of diseases, which necessitates robust diagnostic methods and reliable testing reagents.
Advancements in biotechnology have expanded the possibilities for research and development in microbiology, the integration of molecular biology techniques, as polymerase chain reaction (PCR) and gene sequencing has revolutionized the field by enabling rapid and precise identification of microorganisms. These advancements have paved the way for personalized medicine and targeted therapies, as well as the development of antimicrobial agents to combat drug-resistant strains are all growing demand for microbiology reagents.
The advent of advanced technologies in DNA sequencing, gene-editing, and bioinformatics has revolutionized genomics and molecular biology research, further fueling the demand for microbiology reagents. In addition to these factors, India’s robust growth in contract research and manufacturing services (CRAMS) has further intensified the need for high-quality and reliable reagents. This confluence of factors, stemming from the rapid progress in life sciences and biotechnology, is undeniably propelling the demand for microbiology reagents in India to new heights.
Furthermore, advancements in genomics and proteomics are crucial factors for the expansion of the Indian market. As the trend toward personalized medicine continues to gain momentum, the need for high-quality and reliable microbiology reagents becomes even more crucial. Accurate diagnosis and targeted treatment plans heavily rely on the availability of these reagents, making it imperative for manufacturers to meet the burgeoning demand. Consequently, India has emerged as a hotspot for microbiology reagent manufacturers, many of whom are expanding their operations in the region to cater to the growing needs of healthcare providers and patients alike.
The high cost of reagents, mostly imported, with the dollar parity not working in India’s favor, particularly now that expansion is being seen form semi-urban and rural areas, and rigid guidelines and approval processes for production, import, and use of microbiology reagents by CDSCO and ICMR are seemingly playing spoilsport.
The Western region in India, with its substantial number of diagnostic centers is projected to continue its dominance.
Worldwide too, recent developments in microbiology highlight the industry’s strides toward personalized solutions and innovative technologies. Customized pharmaceuticals, using viral agents like AVV or bacteriophage for unique recombinant drug products, represent a notable advancement. In addition, the growth of advanced therapy medicinal products (ATMPs) necessitates unique quality-control strategies, pushing the field to adopt rapid methodologies and real-time process monitoring. With the industry moving toward personalized medicine, it means that rapid microbiological methods (RMMs) are going to be important for ATMPs as well. Moreover, regulatory changes, especially in endotoxin testing, see a shift toward accepting recombinant reagents globally. Therefore, recent developments showcase a transformative era in microbiology, with a focus on personalized medicine, rapid methods, and global regulatory shifts.
The advent of next-generation sequencing technologies has facilitated the acquisition of large amounts of DNA sequence data at a relatively low cost, leading to numerous breakthroughs in decoding microbial genomes. Among the various genome sequencing activities, metagenomic analysis, which entails the direct analysis of uncultured microbial DNA, has had a profound impact on microbiome research and has emerged as an indispensable technology in this field.
Despite its valuable contributions, metagenomic analysis is a bulk analysis technique that analyzes samples containing a wide diversity of microbes, such as bacteria, yielding information that is averaged across the entire microbial population. In order to gain a deeper understanding of the heterogeneous nature of the microbial world, there is a growing need for single-cell analysis, similar to its use in human cell biology.
With this paradigm shift in mind, comprehensive single-cell genomics technology has become a much-anticipated innovation that is now poised to revolutionize microbiome research. It has the potential to enable the discovery of differences at the strain level and to facilitate a more comprehensive examination of microbial ecosystems.
Successful implementation of this technology is expected to have a profound impact in the field, leading to new discoveries and insights into the diversity and evolution of microbes.
Antimicrobial resistance (AMR) is a global threat to human health and welfare, food safety, and environmental health and, therefore, the rapid detection and quantification of antimicrobial resistance is important. Flow cytometry (FCM) is a powerful analytical tool with a wide range of potential applications in life sciences and biomedicine. The number of FCM applications in the field of microbiology have expanded rapidly, especially in response to AMR challenges. It has been demonstrated that FCM assays can make an important contribution to combating AMR. The development of novel AST frameworks that embed cytometry may inform clinicians in a timely manner.
Additionally, the study of AMR ecology with FCM assays contributes to the understanding of AR acquisition and transmission mechanisms, sources, and routes of the spread of AMR. Future advances involving portable FCM instrumentation, coupled with simpler interpretation software, would support rapid and efficient AMR detection and quantification needed to implement AR monitoring programs and build AMR online databases. These data are critical for agencies and scientists to both manage AMR and assess public health risks.
Advances in rapid microbiology
Rapid microbiology is witnessing significant advancements, transforming the landscape of microbial detection and diagnosis. The constant evolution of technologies is driving the development of innovative methods for quicker and more accurate identification of microorganisms.
The first development revolves around the concept of getting results faster in the field of rapid microbiology. The emphasis is on reducing the turnaround time for microbiological testing without compromising accuracy. This is particularly crucial in scenarios, such as hospital-acquired infections (HAIs), where timely identification of the causative agents is essential for effective intervention and patient care.
One approach to achieving faster results involves the utilization of multiplex real-time polymerase chain reaction (PCR) assays. The development of a new multiplex real-time PCR assay for the rapid screening of hospital-acquired infection agents is a notable breakthrough. This assay allows for the simultaneous detection of multiple pathogens in a single test, expediting the identification process. By targeting specific genetic markers of various microorganisms, this multiplex assay enhances the speed and efficiency of screening for potential infection agents.
The second development underscores the importance of innovative technologies in advancing rapid microbiology. The adoption of novel techniques, such as isothermal amplification, is contributing to faster and simpler microbial detection. Isothermal amplification methods, unlike traditional PCR, do not require thermal cycling, enabling quicker and more energy-efficient DNA amplification.
These advancements in rapid microbiology are crucial in various fields, including clinical diagnostics, food safety, and environmental monitoring. In clinical settings, the ability to swiftly identify pathogens responsible for infections can significantly impact patient outcomes. In food safety, rapid microbiology methods can expedite the detection of foodborne pathogens, ensuring the timely removal of contaminated products from the market. Additionally, in environmental monitoring, quick identification of microbial contaminants is essential for safeguarding public health.
The continuous progress in rapid microbiology reflects the commitment of researchers and industry professionals to address the pressing need for faster and more efficient microbial testing. These developments not only contribute to timely interventions in healthcare settings but also have broader implications for ensuring the safety of food supplies and the environment. As technology continues to advance, the future holds the promise of even more sophisticated and rapid microbiological methods, further shaping the landscape of microbial detection and diagnosis.
Automation reshaping laboratory landscapes
Automation is significantly reshaping the landscape of clinical microbiology laboratories, ushering in transformative changes and offering numerous advantages. Since the inception of the first automated system in 2006, microbiology laboratory automation (MLA) has gained traction across the globe, providing multiple benefits to clinical microbiology laboratories.
The implementation of MLA brings forth several advantages that enhance the efficiency and effectiveness of laboratory operations. One primary benefit is the improvement of workflow by automating labor-intensive and time-consuming processes, such as plate sorting, plate moving, plate streaking, and plate locating. This automation allows for more continuous workflows, extending the working hours of laboratories, ultimately benefiting patient care.
Standardized streaking facilitated by MLA yields a higher number of isolated colonies and ensures uninterrupted incubation processes, thereby enhancing the recovery of pathogens from culture. Additionally, MLA contributes to reducing human errors associated with manual tasks. Furthermore, the elimination of repetitive tasks for laboratory staff helps mitigate ergonomic issues.
Another notable strength of MLA is its ability to provide reproducible results with image documentation. When coupled with barcoding capability, digital imaging ensures traceability, offering significant advantages to laboratories.
Economically, the improvement in laboratory efficiency through MLA cannot be overlooked, especially in the face of increasing testing demands and staffing shortages in microbiology laboratories. However, the high initial costs associated with procuring and installing MLA systems need to be carefully considered.
Despite the evident benefits, the installation of MLA systems comes with its set of challenges, including the risk of downtime and the necessity for a backup plan. However, it is essential to note that MLA systems are modular, allowing for individualized levels of automation and a gradual transition, mitigating potential disruptions.
At the simplest level, laboratory automation exists to remove the more repetitive tasks, such as performing dilutions or plate spotting. At a more advanced level, entire workflows can be automated, as with the case of robotics designed for bacterial endotoxin testing. In between are automated workflows to manage complex tests, balance multiple workflows, and reduce the most time-consuming steps, such as sample preparation.
Three current examples in the microbiology laboratory are:
Reproducibility is a common concern in microbiology, especially for techniques reliant upon the variances of laboratory analysts. Examples of automation technologies that can drive improvements include robotics for streaking samples onto the agar plates and inoculating liquid culture media. These forms of laboratory automation can enable a greater rate of experimental data capture, increased volume of results, and the use of a wider range of controls.
Sample traceability is important for business efficiency, and it is a core part of cGMP. The primary way traceability is automated is with the bar-coding of samples. Automated solutions connect bar code scanners, printers, plate readers, incubators, test devices, and information management solutions.
Data accuracy is essential, not only to meet cGMP requirements but also to ensure that appropriate insights are drawn. A new array of automated processes is incorporating machine learning algorithms that strengthen the insights from data evaluation. These technologies become more sophisticated by learning from large pools of real samples.
Future trends and challenges
In terms of automation advances and what the microbiology laboratory of the future is set to exploit, five key opportunity areas are identified.
- Biosensors. In the areas of drug development and disease diagnosis, lab-on-a-chip technology holds the potential to move laboratory analyses away from traditional diagnostic platforms to more powerful technology. Biosensor devices consist of small chip-based miniaturized platforms that can be used for the immediate detection of different organisms and to assess how they interact with different compounds, by using analytes of low volumes. These devices are based on microfluidic technologies. Microfluidic devices work by assessing the physical and chemical properties of liquids at a microscale, enabling dozens of tests to be conducted simultaneously.
A related development has seen the creation of a microfluidic fluorescence quantitative PCR system with a pneumatic valve and a tree structure that was developed by using 3D printing technology. This is necessary because the objective is to create well-defined microenvironments that mimic the natural habitats of bacteria.
- Assessing microbial colonies early. The majority of methods within the microbiology laboratory are growth-based, reliant upon agar or broth culture media. It is often the case that obtaining a result as fast as possible aids in faster reporting and release of product. Several technologies are focused on providing a faster reading of agar plates. Examples include the use of light forward-scattering and learning algorithms and digital imaging of cellular autofluorescence to detect and enumerate growing microcolonies many generations before they become visible to the eye.
Where multiple incubation steps are required, some systems have the ability to automatically transfer culture media to specialized incubators operating at different temperature ranges through robotic systems.
An alternative option is the array of automated colony counting technologies available. Some of the technologies have struggled to accurately count mixed cultures, although improvements have been made with innovations like hyperspectral imaging technology.
- Microbial identification. MALDI-TOF has become the identification method of choice in many laboratories, and it has drastically cut down preparation time and time-to-result. In terms of future developments with identification technology, there is considerable interest in surface-enhanced Raman spectroscopy (SERS) analysis. This is used in conjunction with a deep learning model, based on optimizing the route to classify the signals of two common bacteria and their resident media without the need for any separation procedures. This form of Raman spectroscopy functions by sending light through a sample to observe how it scatters. Current developments indicate it is possible to classify various bacteria in different media with accuracies of up to 98 percent. The algorithm is based on the functionality of a dual-branch wide-kernel network and it is trained to reveal and assess structural information about a given sample (the spectral fingerprint).
There is an associated benefit in relation to drug development, where convolutional neural networks (CNN) can be trained to classify noisy bacterial spectra by isolate, empiric treatment, and drug resistance. Hence, such technology can assist with developing susceptibility profiles.
- Transfer of data to LIMS or equivalent systems. To reduce data errors, and to speed up the analysis process, the smart microbiology laboratory will seek to connect as many instruments to laboratory information management systems (LIMS) as possible. As microbiology equipment progresses, the required interfaces between the equipment and LIMS is made possible. One example is the connection of microbial identification systems like MALDI-TOF MS into these workflows.
To accommodate advances in laboratory analytical equipment and to enable fuller insights to be gained from data, LIMS technology needs to advance. LIMS is often hampered by limited database flexibility owing to instrumentation, insufficient traceability, inadequate data protection, and low management efficiency. These obstacles are being overcome by projects seeking to construct smart central management systems, modelled on the human brain based on the LIMS; that is combined with current advanced technologies, such as information modelling and artificial intelligence (AI).
Many of the elements of laboratory automation can support this next-generation LIMS, including high-definition cameras, sensors, automatic control units, and robotic arms.
- Use of AI to analyze data. AI has the potential to analyze Big Data and find patterns and insights that could enhance understanding microbial growth, improve microbial identification, enable using microorganisms to produce new compounds in biotechnological applications, and assist with antimicrobial drug development. In each of these cases, processing data becomes much faster, and more meaningful patterns can be captured. Giving any individual many different data points without synthesizing and analyzing the information makes it difficult to comprehend the information. AI can conduct the analysis and provide real insights, direction, and even advice. It is likely that developments within the pathology sector will provide the basis for pharma. The FDA has approved a digital pathology system, an AI platform called OsteoDetect, and a software-as-a-service (SaaS) application, Glooko, both of which use AI to advise on results outcomes.
AI can also support existing laboratory procedures. For example, it can serve as a supplementary or a validation tool in imaging analytics and help process more slides or culture plates in a shorter duration.
New frontiers in microbiology
The convergence of nanotechnology and microbiology opens new avenues for more sensitive and efficient diagnostics. Utilizing nanomaterials enhances the sensitivity and specificity of detection methods, allowing for rapid and accurate identification of microorganisms. This trend signifies a move toward more precise and targeted diagnostics, reducing false positives and negatives. Recently, the development of atmospheric pressure matrix-assisted desorption/ionization mass spectrometry (AP MALDI MS) has made contributions not only to biomolecule analysis but also to spatial distribution. AP MALDI works as a powerful tool in multiple domains as compared to conventional MALDI MS and has attracted the attention of scientists.
The future of automated microbiology laboratories requires collaborative efforts among researchers, industry stakeholders, and regulatory bodies. Collaborative initiatives can drive innovation, facilitate knowledge exchange, and address challenges collectively. Establishing a framework for collaboration ensures that advancements in automated microbiology are robust, validated, and aligned with the needs of diverse applications, from clinical diagnostics to industrial quality control.
Way forward
The future of microbiology is poised for groundbreaking transformations, driven by the convergence of cutting-edge technologies and innovative research endeavors. Automation, a central force reshaping laboratory landscapes, is expected to play a pivotal role in the microbiology of the future. The integration of AI, robotics, and advanced data analytics promises to create highly automated microbiology labs, revolutionizing sample processing, testing, and data analysis. This leap toward automation not only enhances efficiency but also ensures the reliability of microbiological results by minimizing human errors.
Despite all exciting prospects, challenges in data management and seamless integration require collaborative efforts. As technology evolves, the microbiology landscape is set to deliver more precise diagnostics, accelerated detection methods, and sophisticated automated systems, ushering in a new era of transformative capabilities in microbial research and diagnostics.