DNA Sequencers
Emerging technologies to revolutionize genome analysis

Sequencing capabilities today are work-in-progress; they are a long way off to where sequencing becomes ubiquitous, comprehensive, and accurate.
Forty years ago, the advent of Sanger sequencing was revolutionary as it allowed complete genome sequences to be deciphered for the first time. A second revolution came when next-generation sequencing (NGS) technologies appeared, which made genome sequencing much cheaper and faster. In particular, short-read sequencing approaches, including sequencing by synthesis, ion-semiconductor sequencing, and nano-ball sequencing have dramatically enhanced these outputs. Third-generation long-read sequencing is a step forward, allowing overcoming certain limitations of short-read sequencings, such as reliability to solve repeated sequences and large genomic rearrangements. The combination of adjacent technologies, such as nanotechnology – nanopore, and complementary methods, including in-situ nucleic-acid sequencing, and microscopy-based sequencing, was crucial to driving massively parallel DNA sequencing. This facilitated the introduction of NGS technologies to unveil complex biological contexts around disease mechanisms. Their utilization for genetic diagnosis purposes is currently receiving extraordinary attention.
Demand for accurate, inexpensive, and fast DNA sequencing data has led to the evolution and dominance of a new generation of sequencing technologies. In addition, different sciences are receiving benefits of this technique, including forensic sciences, molecular biology, biotechnology, genetics, anthropology, and archaeology. DNA sequencers are expected to revolutionize the conceptual foundation of these fields.
Researchers are now using innovative technologies for genome mapping that take lesser time for completion. Other than this, the reduction in sequencing costs has led many companies to invest in R&D activities, and offer cost-effective solutions, such as whole genome sequencing, de-novo sequencing, and specific disease diagnosis. The demand for DNA sequencers has increased significantly with surge in sequencing applications and rise in technological advancements in DNA sequencing. In addition, surge in the number of genome-mapping programs globally and increase in R&D investment is expected to drive the market growth in the years to come.
The global DNA sequencing market is expected to grow by USD 17.92 billion during 2021–2025. This marks a significant market slowdown compared to the 2020 growth estimates due to the impact of the COVID-19 pandemic in the first half of 2020. However, healthy growth is expected to continue over the next five years, and the market is expected to grow at a CAGR of over 19 percent. Among the product segment, consumables are expected to dominate the overall market. The wide availability of reagents and kits to cater to all steps of library construction, such as DNA fragmentation, enrichment, adapter ligation, amplification, and quality control contributes to a larger share of the segment. The third-generation sequencing methods, such as nanopore and single-molecule real-time sequencing, are projected to witness a considerable CAGR in coming years.
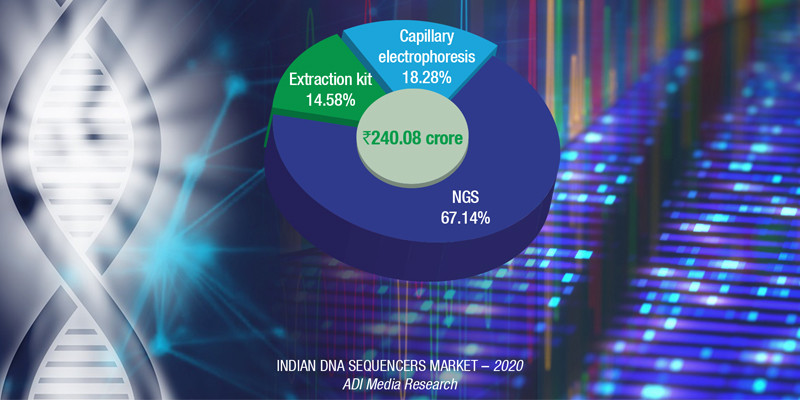
The Indian DNA sequencers market saw a sharp drop in 2020, of 31.5 percent. With research and clinical institutes under lockdown, the technicians were moved to conducting the PCR tests.
What really took off was RNA extraction. And this trend is expected to stay. Several protocols for RNA extraction and RT-qPCR that are simpler and less expensive than prevailing methods are possible. First, isopropanol precipitation is shown to provide an effective means of RNA extraction from nasopharyngeal (NP) swab samples. Second, direct addition of NP swab samples to RT-qPCRs is evaluated without an RNA-extraction step. A simple, inexpensive swab collection solution suitable for direct addition is validated using contrived swab samples. Third, an open-source master mix for RT-qPCR is described that permits detection of viral RNA in NP swab samples, with a limit of detection of approximately 50 RNA copies per reaction. Quantification cycle (Cq) values for purified RNA from 30 known positive clinical samples showed a strong correlation (r2=0.98) between this homemade master-mix and commercial TaqPath master-mix. Lastly, end-point fluorescence imaging is found to provide an accurate diagnostic readout without requiring a qPCR thermocycler. Adoption of these simple, open-source methods has the potential to reduce the time and expense of COVID-19 testing.
COVID-19 has drawn attention to the relevance of sequencing and pathogen surveillance. If there is one tool in the COVID-19 pandemic response, which India has been slow in adoption and has used sub-optimally, it is genome sequencing. The scale of exercise has been dismal. To cite data, less than 1 percent of the total positive samples since January 2021 through March 18, 2021, have been sequenced.
Genome sequencing is an exercise that studies changes in the structure of the virus over time. A combination of changes (mutations) in the ribonucleic acid of the virus can give birth to a new variant. The exercise is carried out only on samples that have tested positive for the virus.
Other countries kicked off the exercise right at the beginning of the pandemic. In fact, many of its neighbors in the South and Southeast Asia have re-embarked on a remarkable effort to quickly scale up their very limited sequencing capacities to identify whether India’s epidemic – fueled by variants such as B.1.617 – is spilling over into their communities, or if their outbreaks have origins elsewhere. In some cases, they are working around the clock and with insufficient resources. Specifically, they want to know whether worrying variants are circulating, so that they can assess the risks. Identifying worrying variants will help governments to make decisions about responses and restrictions, and whether more aggressive interventions are needed. Given India’s significant caseload, it should have jumped on to the bandwagon quickly as well.
It is expected that the government will allocate more than the current 0.8 percent of GDP for this segment. In any case, the Department of Biotechnology and the Ministry of Health are looking at how to involve the private sector. Discussions are on with certain major corporate hospitals and private labs that are into sequencing. The government cannot fund them because the amount of money that is required is huge. A framework is being developed where private entities can do sequencing, but all sequences should finally find one repository in terms of data repository so that the whole nation’s picture is reflected.
Genomics is also important for other epidemic diseases like dengue, chikungunya, and influenza because once COVID goes down, these will be back. Monitoring for variants and mutants is also very important. Emerging variants, with evidence of higher transmissibility and immune escape, demand re-strategized responses.
Sequencing capabilities today are work-in-progress; they are a long way off to where sequencing becomes ubiquitous, comprehensive, and accurate. Despite the huge cost reductions (in 2007 it cost USD 300,000 to sequence a human genome, compared to USD 600 today), cost is still the biggest barrier to wider adoption.
It is highly likely that the industry will have a blood-based cancer test in the next 5 to 10 years that will be able to detect the early presence of cancer, and that will be routinely used to monitor health. This is likely for other diseases too. Genome sequencing is headed to be a part of regular medical check-ups, where mutations and epigenetic changes can be tracked through life. But for these advancements the market may need to wait a little longer, until the ability to interpret the human genome catches up with the data that sequencing technologies can already provide.